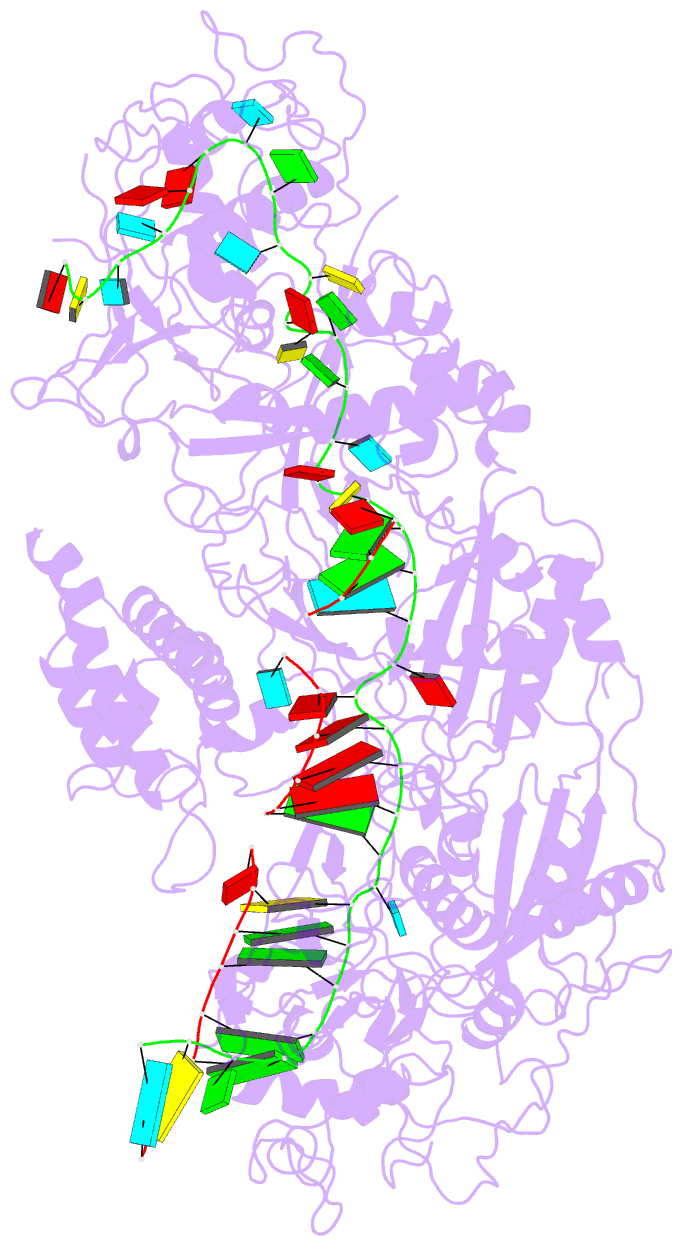
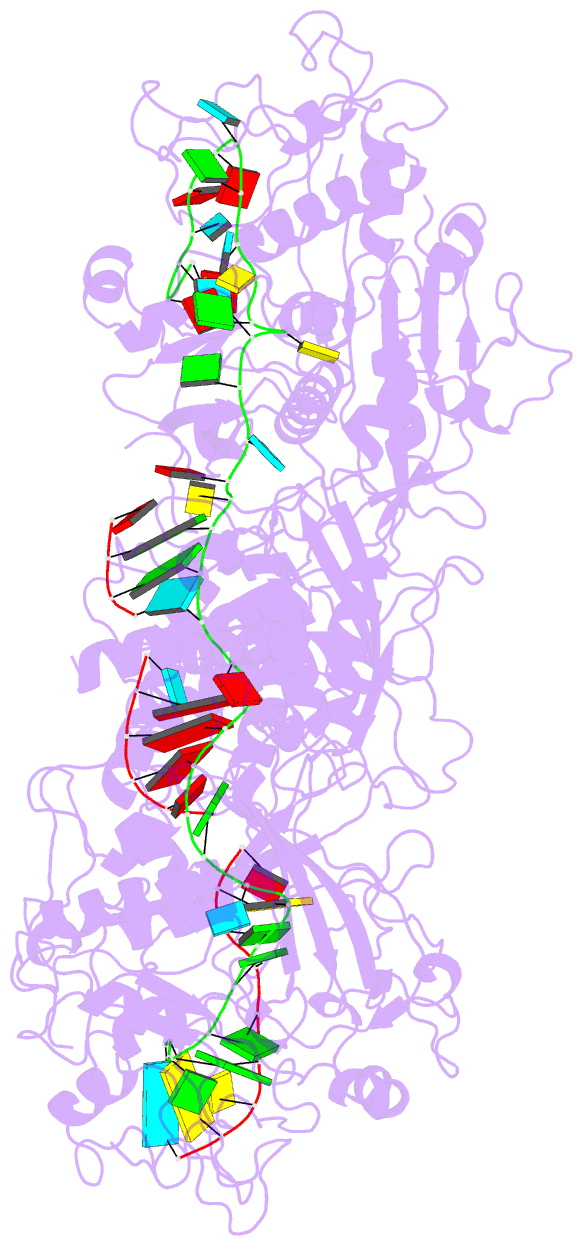
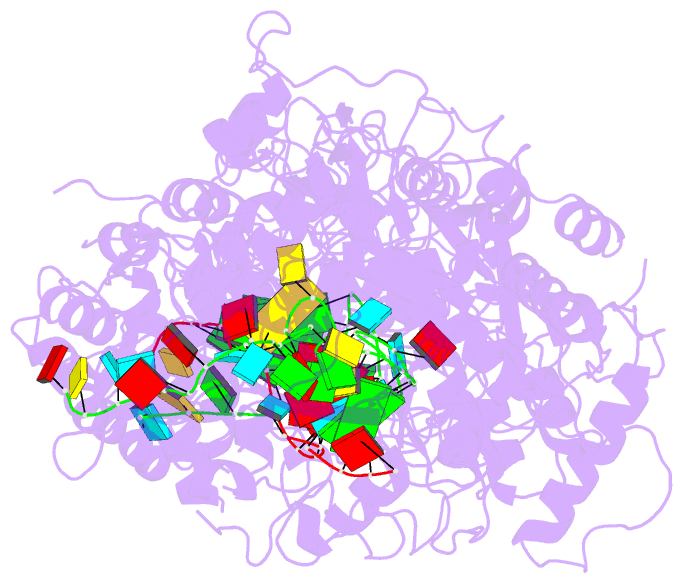
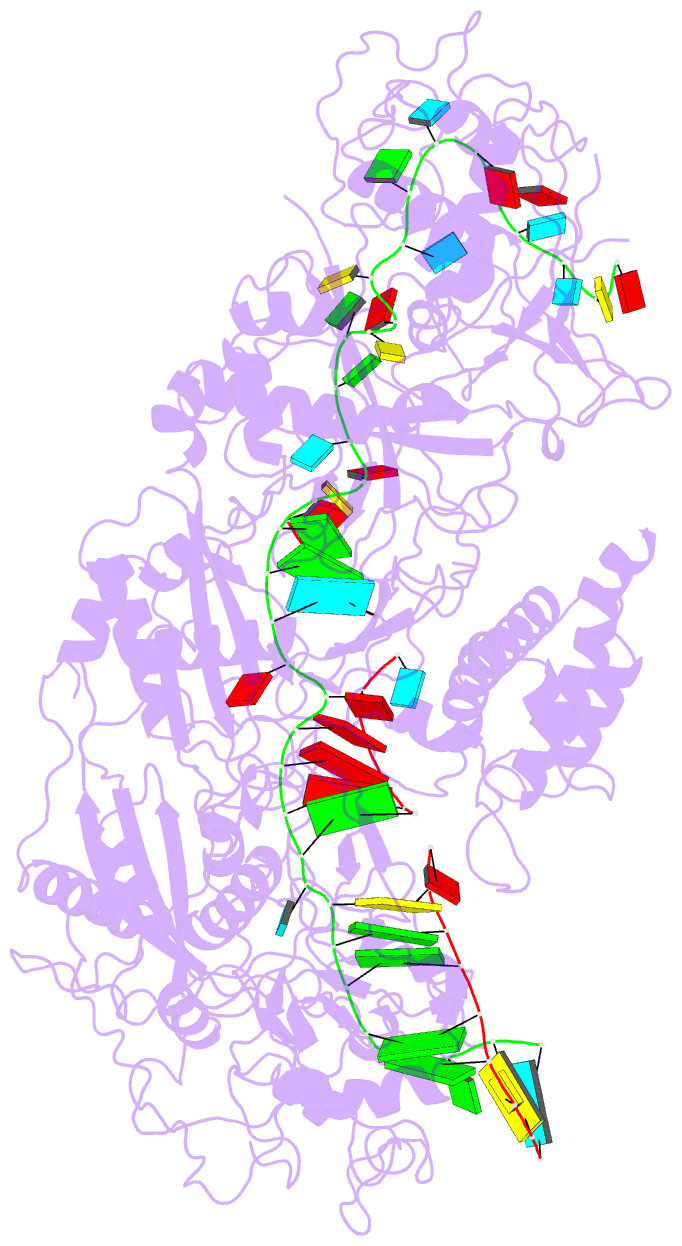
Summary information and primary citation
- PDB-id
- 8d9i; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- cryo-EM (3.62 Å)
- Summary
- Gramp non-matching pfs-with mg
- Reference
- Hu C, van Beljouw SPB, Nam KH, Schuler G, Ding F, Cui Y, Rodriguez-Molina A, Haagsma AC, Valk M, Pabst M, Brouns SJJ, Ke A (2022): "Craspase is a CRISPR RNA-guided, RNA-activated protease." Science, 377, 1278-1285. doi: 10.1126/science.add5064.
- Abstract
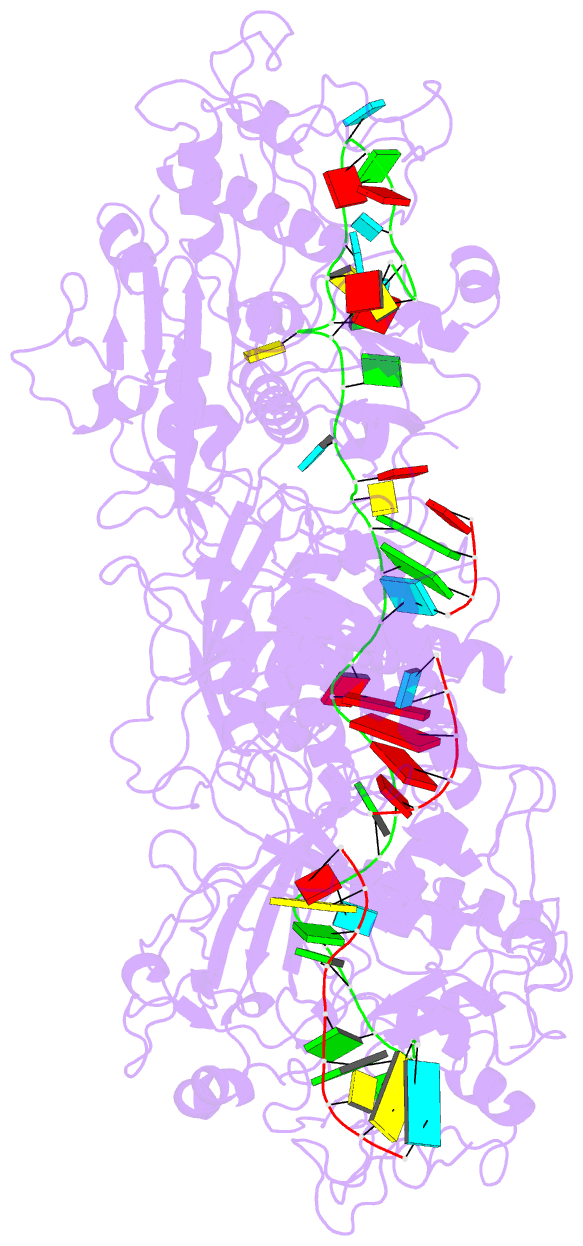
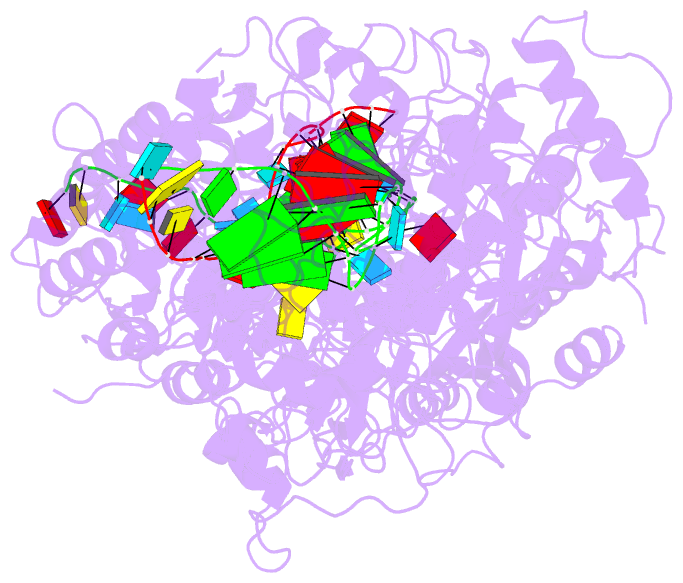
- The Type III-E RNA-targeting effector complex (gRAMP/Cas7-11) is associated with a caspase-like protein (TPR-CHAT/Csx29) to form Craspase (CRISPR-guided caspase). Here we use cryo-electron microscopy snapshots of Craspase to explain its target RNA cleavage and protease activation mechanisms. Target-guide pairing extending into the 5' region of the guide RNA displaces a gating loop in gRAMP, which triggers an extensive conformational relay that allosterically aligns the protease catalytic dyad and opens an amino acid sidechain-binding pocket. We further define Csx30 as the endogenous protein substrate that is site-specifically proteolyzed by RNA-activated Craspase. This protease activity is switched off by target RNA cleavage by gRAMP, and is not activated by RNA targets containing a matching protospacer flanking sequence. We thus conclude that Craspase is a target RNA-activated protease with self-regulatory capacity.