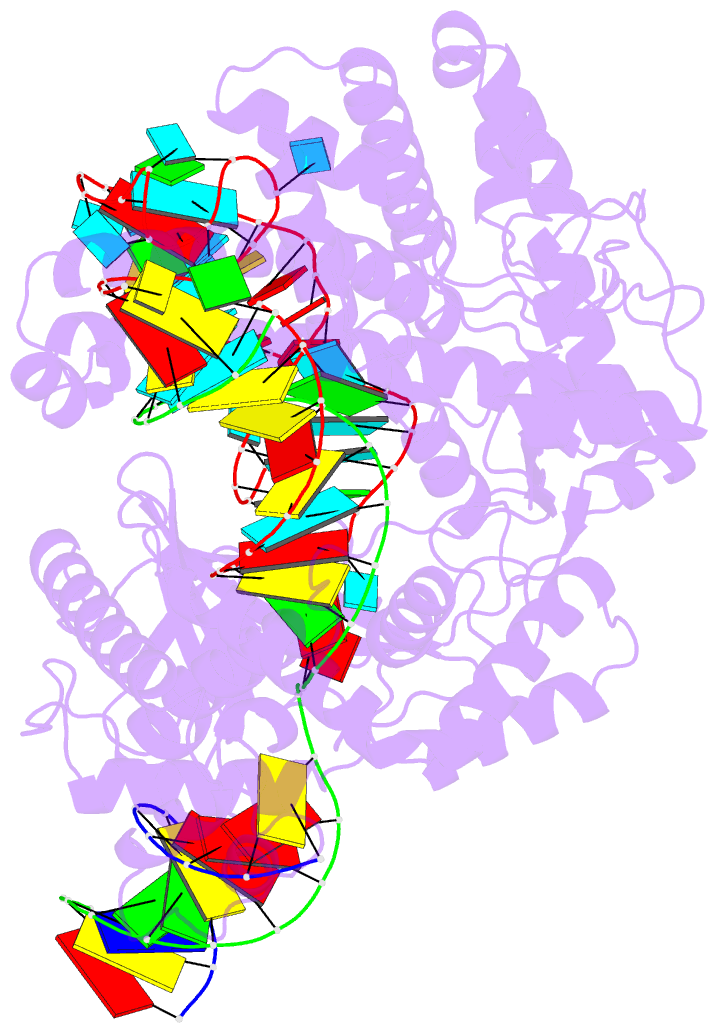
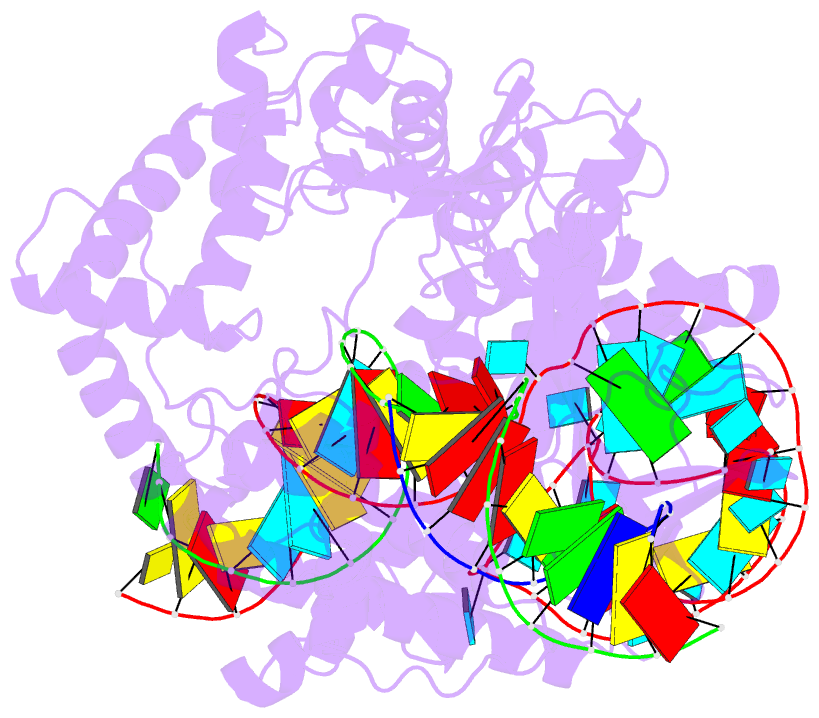
Summary information and primary citation
- PDB-id
- 8dc2; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA-DNA
- Method
- cryo-EM (2.99 Å)
- Summary
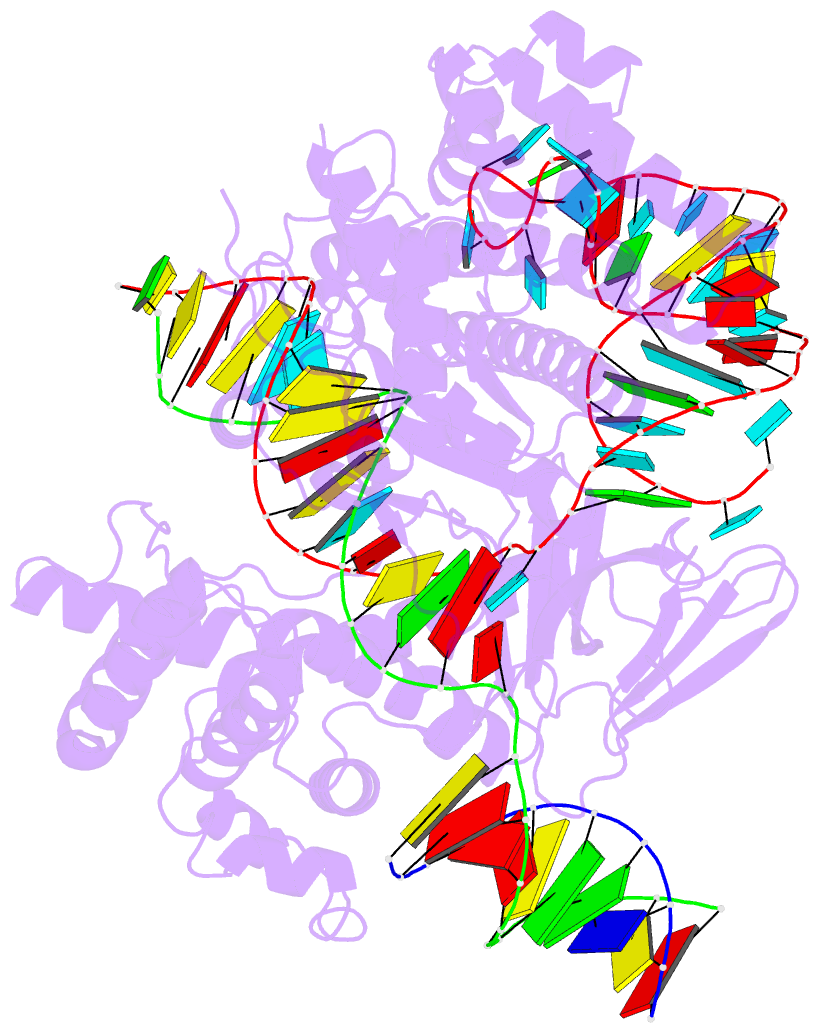
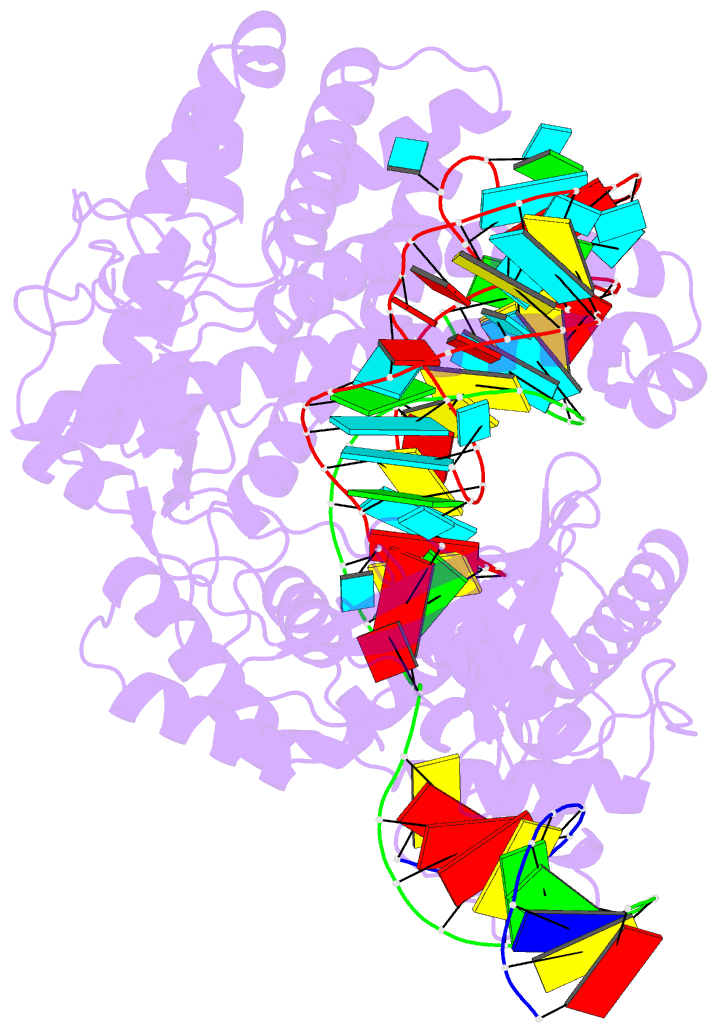
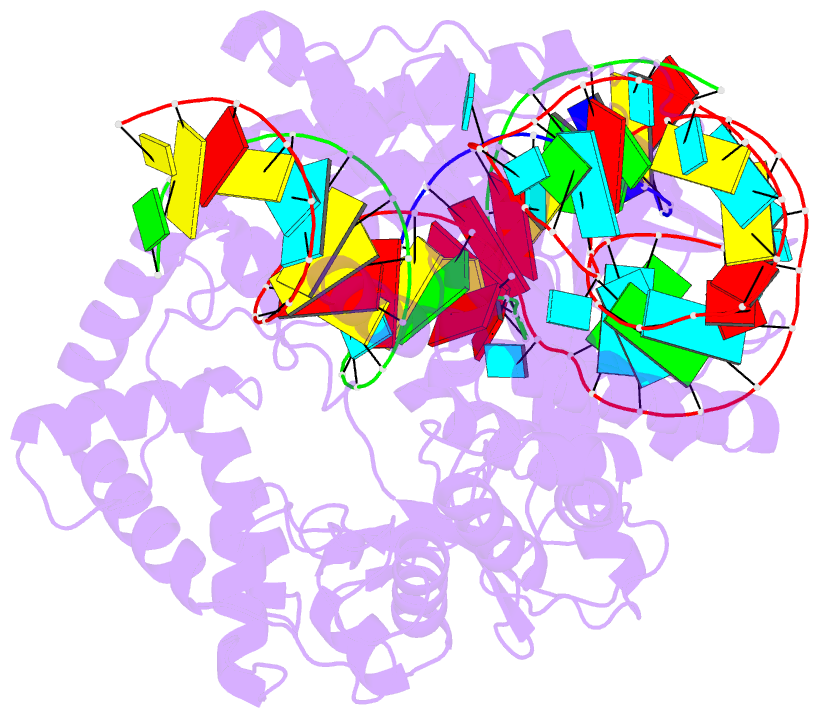
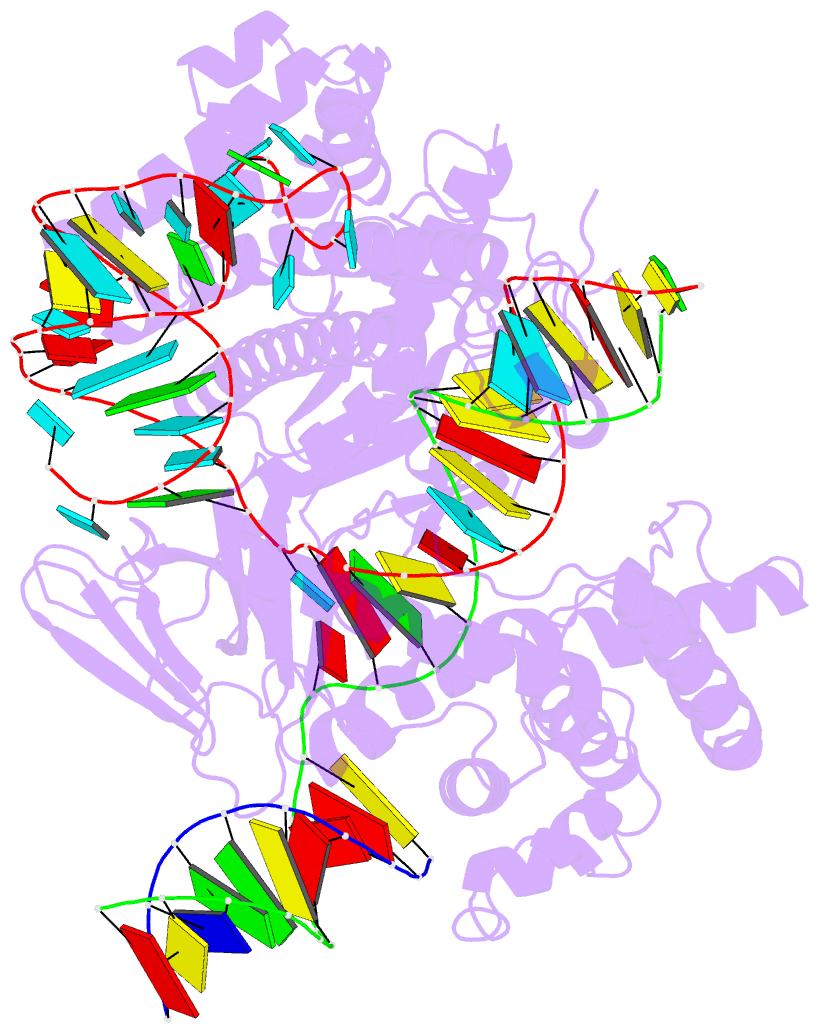
- cryo-EM structure of caslambda (cas12l) bound to crrna and DNA
- Reference
- Al-Shayeb B, Skopintsev P, Soczek KM, Stahl EC, Li Z, Groover E, Smock D, Eggers AR, Pausch P, Cress BF, Huang CJ, Staskawicz B, Savage DF, Jacobsen SE, Banfield JF, Doudna JA (2022): "Diverse virus-encoded CRISPR-Cas systems include streamlined genome editors." Cell, 185, 4574-4586.e16. doi: 10.1016/j.cell.2022.10.020.
- Abstract
- CRISPR-Cas systems are host-encoded pathways that protect microbes from viral infection using an adaptive RNA-guided mechanism. Using genome-resolved metagenomics, we find that CRISPR systems are also encoded in diverse bacteriophages, where they occur as divergent and hypercompact anti-viral systems. Bacteriophage-encoded CRISPR systems belong to all six known CRISPR-Cas types, though some lack crucial components, suggesting alternate functional roles or host complementation. We describe multiple new Cas9-like proteins and 44 families related to type V CRISPR-Cas systems, including the Casλ RNA-guided nuclease family. Among the most divergent of the new enzymes identified, Casλ recognizes double-stranded DNA using a uniquely structured CRISPR RNA (crRNA). The Casλ-RNA-DNA structure determined by cryoelectron microscopy reveals a compact bilobed architecture capable of inducing genome editing in mammalian, Arabidopsis, and hexaploid wheat cells. These findings reveal a new source of CRISPR-Cas enzymes in phages and highlight their value as genome editors in plant and human cells.