Summary information and primary citation
- PDB-id
- 8dp3; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- immune system-RNA
- Method
- X-ray (1.91 Å)
- Summary
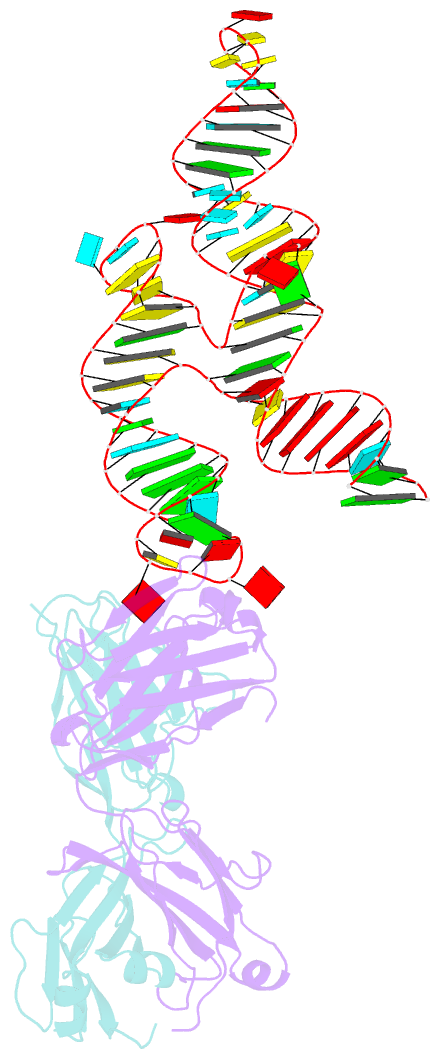
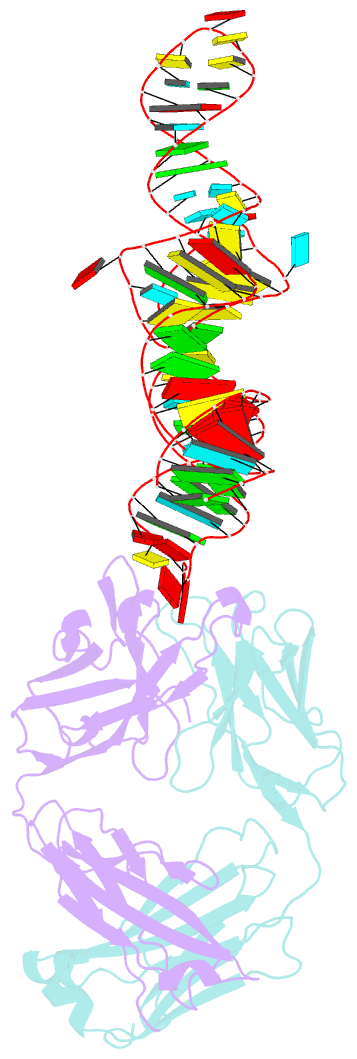
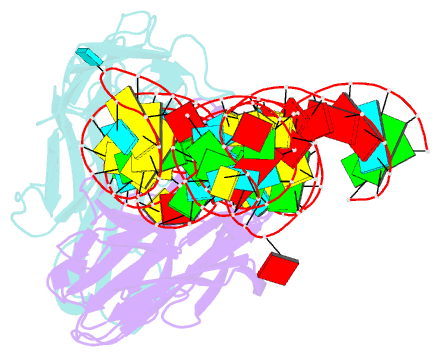
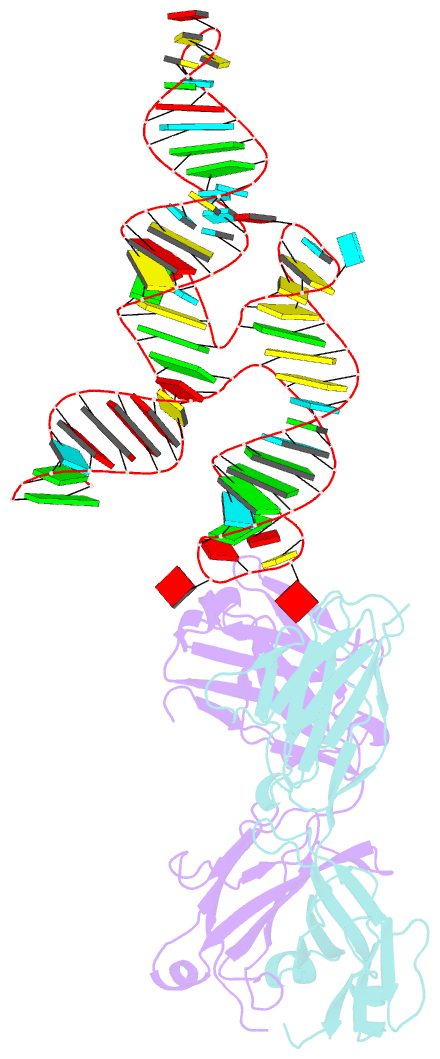
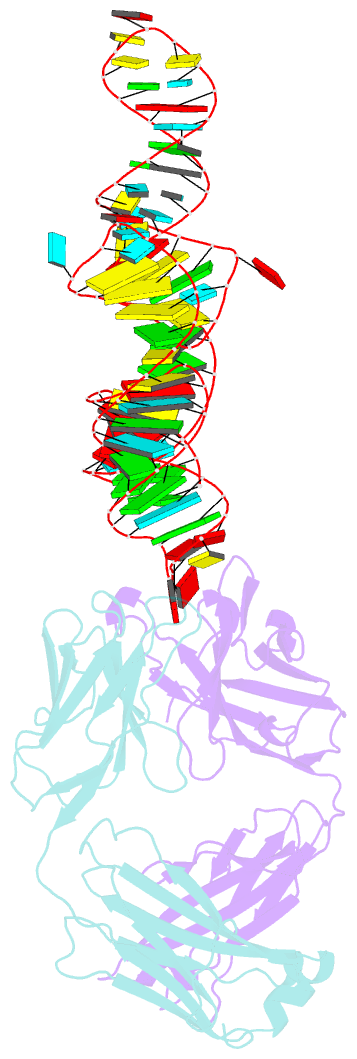
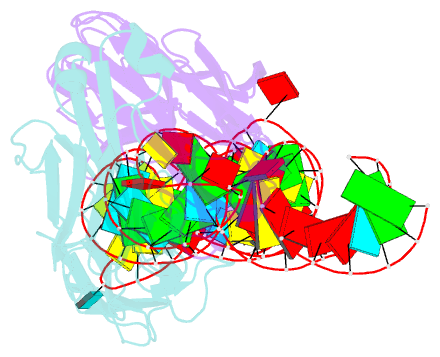
- Crystal structure of coxsackievirus b3 cloverleaf RNA replication element
- Reference
- Das NK, Hollmann NM, Vogt J, Sevdalis SE, Banna HA, Ojha M, Koirala D (2023): "Crystal structure of a highly conserved enteroviral 5' cloverleaf RNA replication element." Nat Commun, 14, 1955. doi: 10.1038/s41467-023-37658-8.
- Abstract
- The extreme 5'-end of the enterovirus RNA genome contains a conserved cloverleaf-like domain that recruits 3CD and PCBP proteins required for initiating genome replication. Here, we report the crystal structure at 1.9 Å resolution of this domain from the CVB3 genome in complex with an antibody chaperone. The RNA folds into an antiparallel H-type four-way junction comprising four subdomains with co-axially stacked sA-sD and sB-sC helices. Long-range interactions between a conserved A40 in the sC-loop and Py-Py helix within the sD subdomain organize near-parallel orientations of the sA-sB and sC-sD helices. Our NMR studies confirm that these long-range interactions occur in solution and without the chaperone. The phylogenetic analyses indicate that our crystal structure represents a conserved architecture of enteroviral cloverleaf-like domains, including the A40 and Py-Py interactions. The protein binding studies further suggest that the H-shape architecture provides a ready-made platform to recruit 3CD and PCBP2 for viral replication.