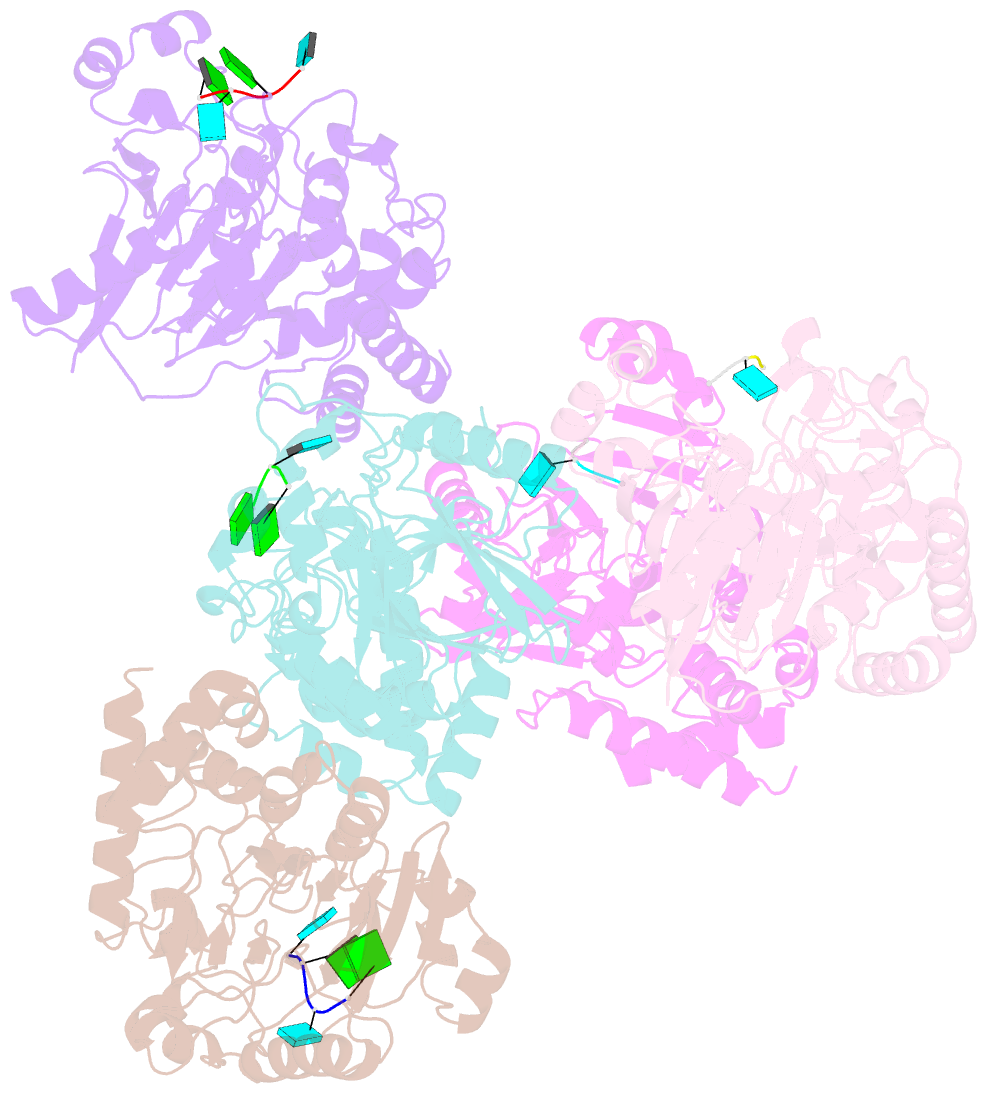
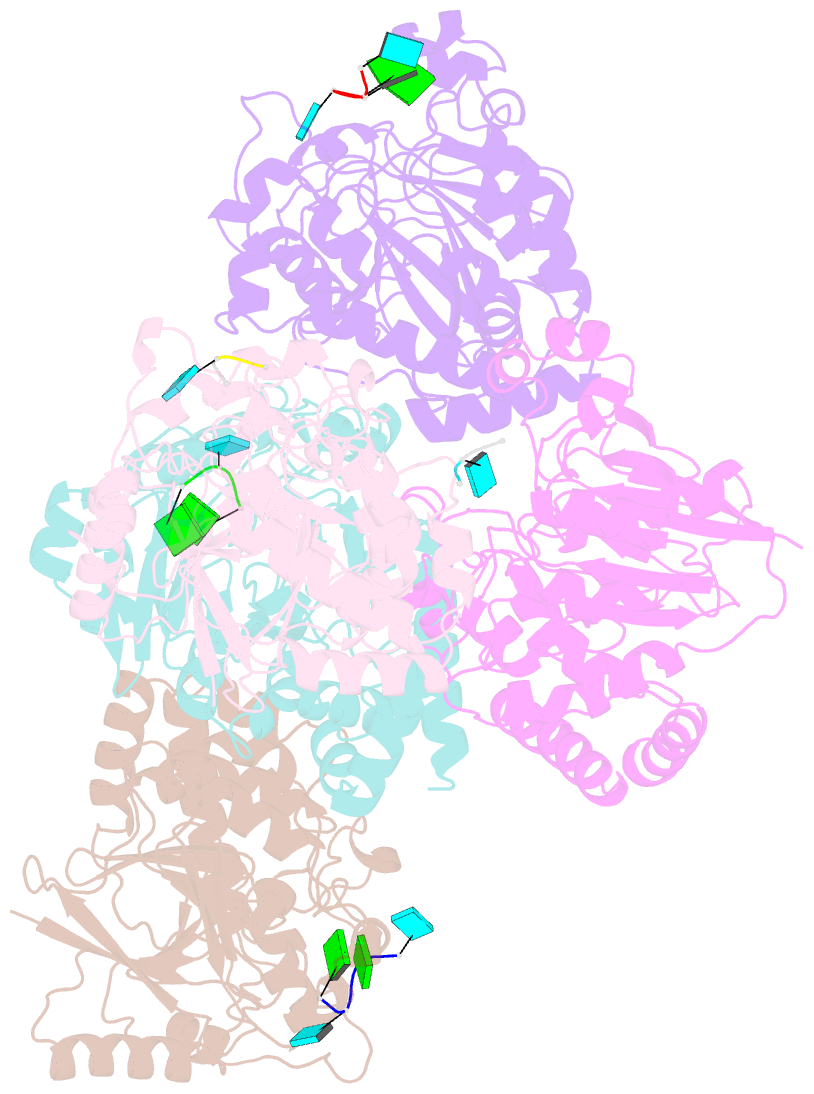
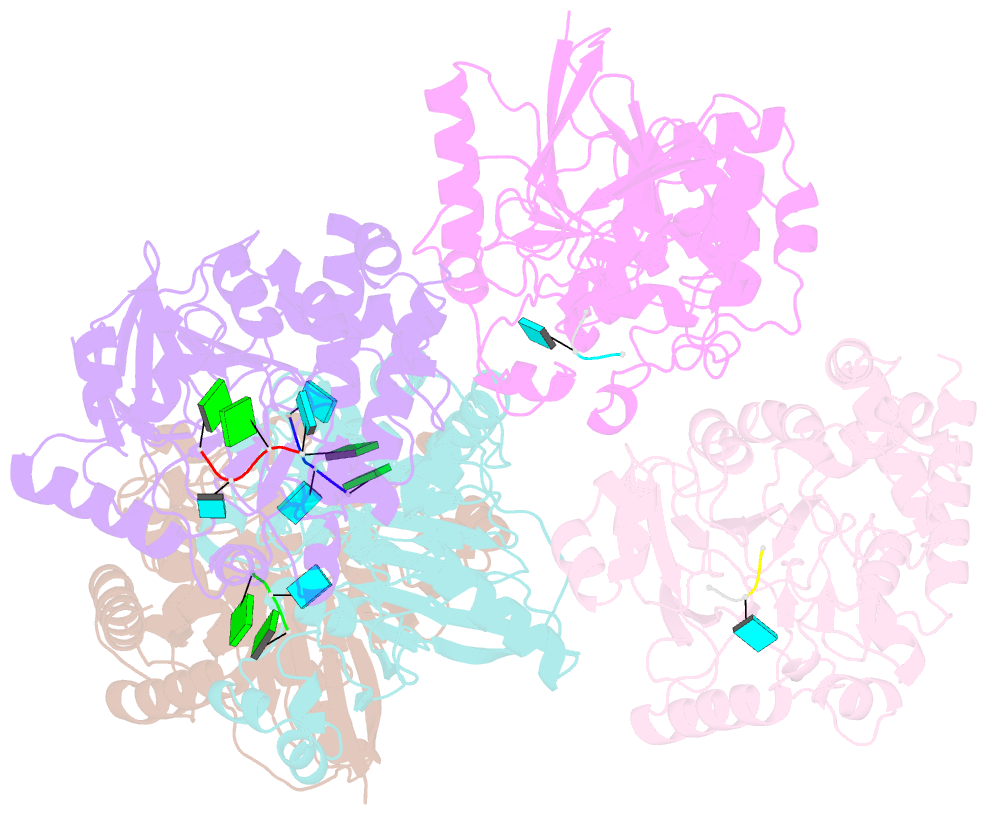
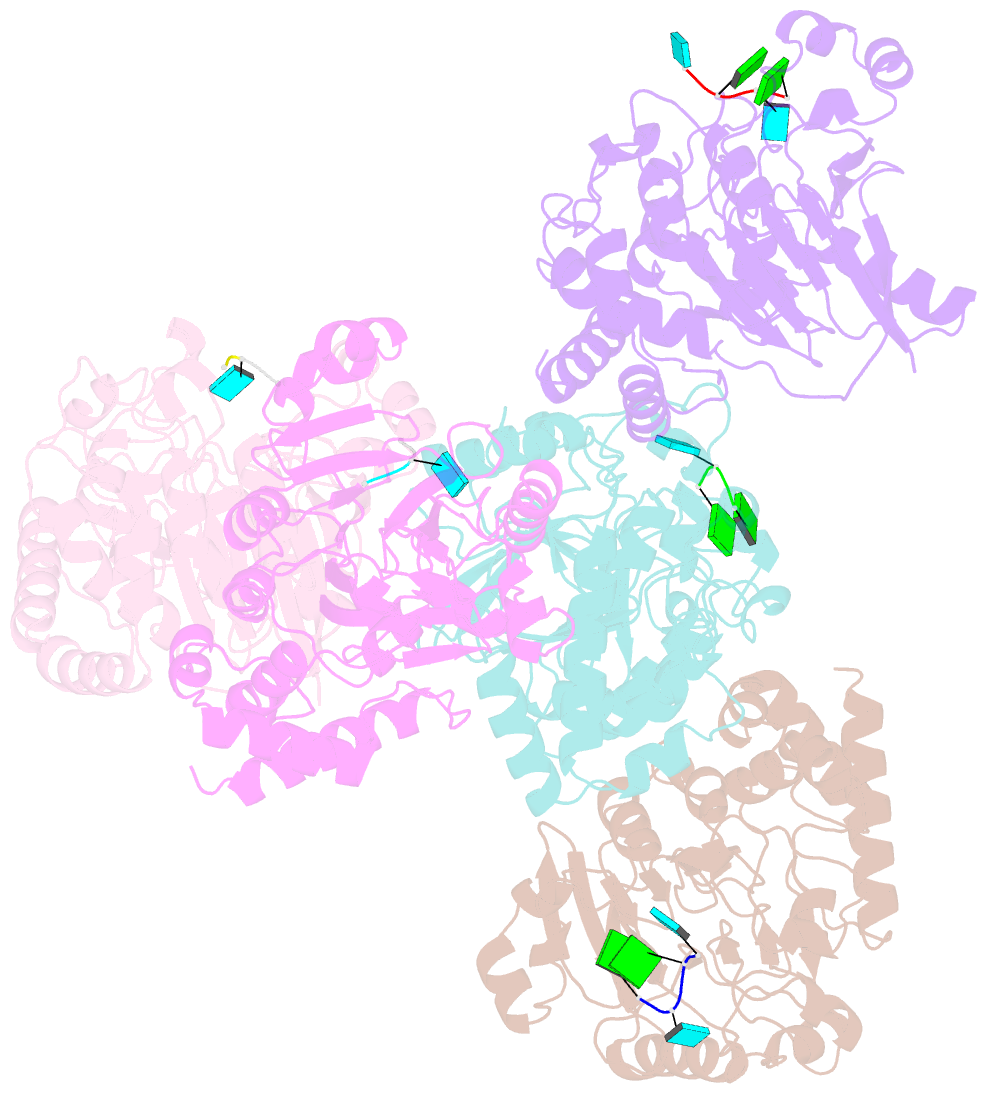
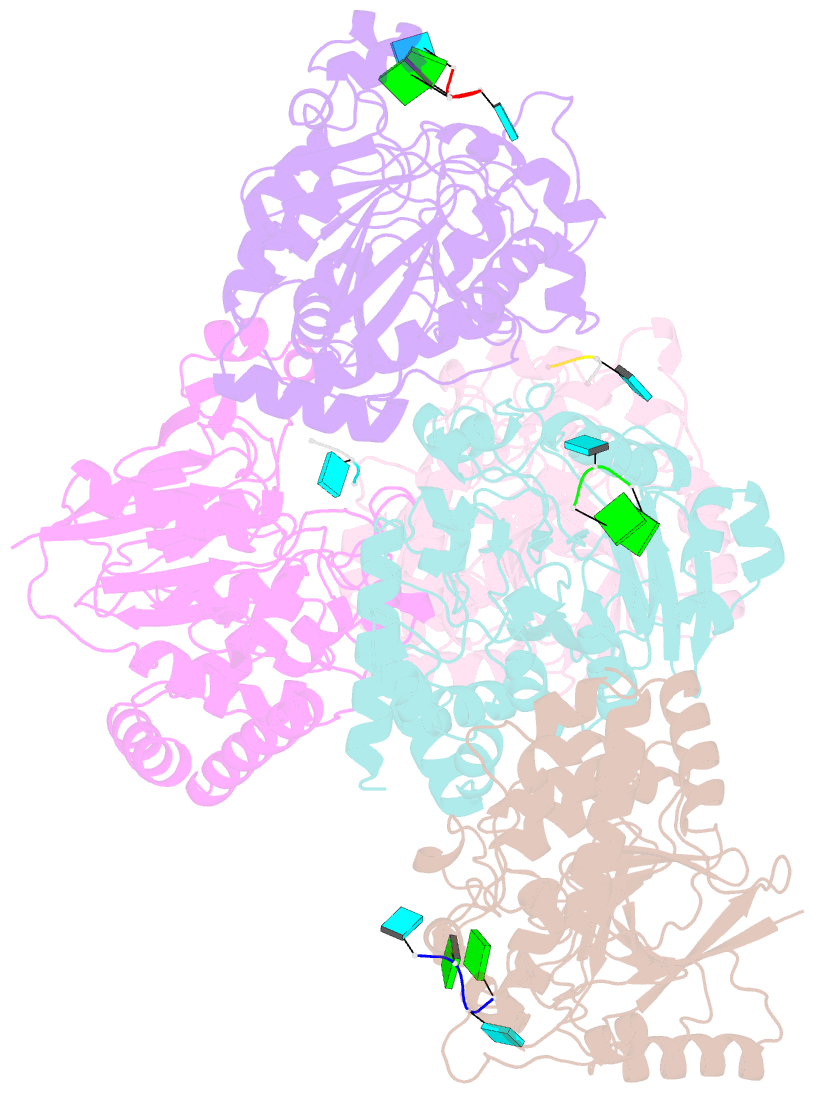
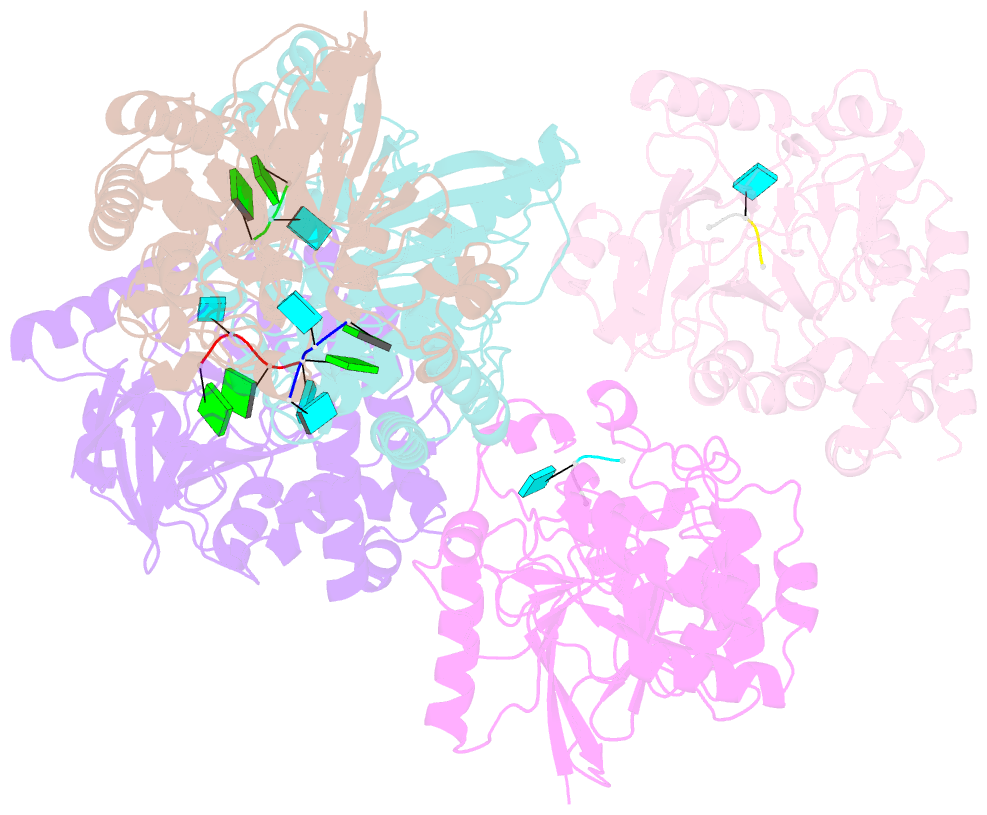
Summary information and primary citation
- PDB-id
- 8dzk; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-RNA
- Method
- X-ray (2.1 Å)
- Summary
- Dbr1 in complex with 5-mer cleavage product
- Reference
- Clark NE, Katolik A, Welch A, Schorl C, Holloway SP, Schuermann JP, Hart PJ, Taylor AB, Damha MJ, Fairbrother WG (2022): "Crystal Structure of the RNA Lariat Debranching Enzyme Dbr1 with Hydrolyzed Phosphorothioate RNA Product." Biochemistry, 61, 2933-2939. doi: 10.1021/acs.biochem.2c00590.
- Abstract
- The RNA lariat debranching enzyme is the sole enzyme responsible for hydrolyzing the 2'-5' phosphodiester bond in RNA lariats produced by the spliceosome. Here, we test the ability of Dbr1 to hydrolyze branched RNAs (bRNAs) that contain a 2'-5'-phosphorothioate linkage, a modification commonly used to resist degradation. We attempted to cocrystallize a phosphorothioate-branched RNA (PS-bRNA) with wild-type Entamoeba histolytica Dbr1 (EhDbr1) but observed in-crystal hydrolysis of the phosphorothioate bond. The crystal structure revealed EhDbr1 in a product-bound state, with the hydrolyzed 2'-5' fragment of the PS-bRNA mimicking the binding mode of the native bRNA substrate. These findings suggest that product inhibition may contribute to the kinetic mechanism of Dbr1. We show that Dbr1 enzymes cleave phosphorothioate linkages at rates ∼10,000-fold more slowly than native phosphate linkages. This new product-bound crystal structure offers atomic details, which can aid inhibitor design. Dbr1 inhibitors could be therapeutic or investigative compounds for human diseases such as human immunodeficiency virus (HIV), amyotrophic lateral sclerosis (ALS), cancer, and viral encephalitis.