Summary information and primary citation
- PDB-id
- 8e2l; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- replication-DNA
- Method
- cryo-EM (3.51 Å)
- Summary
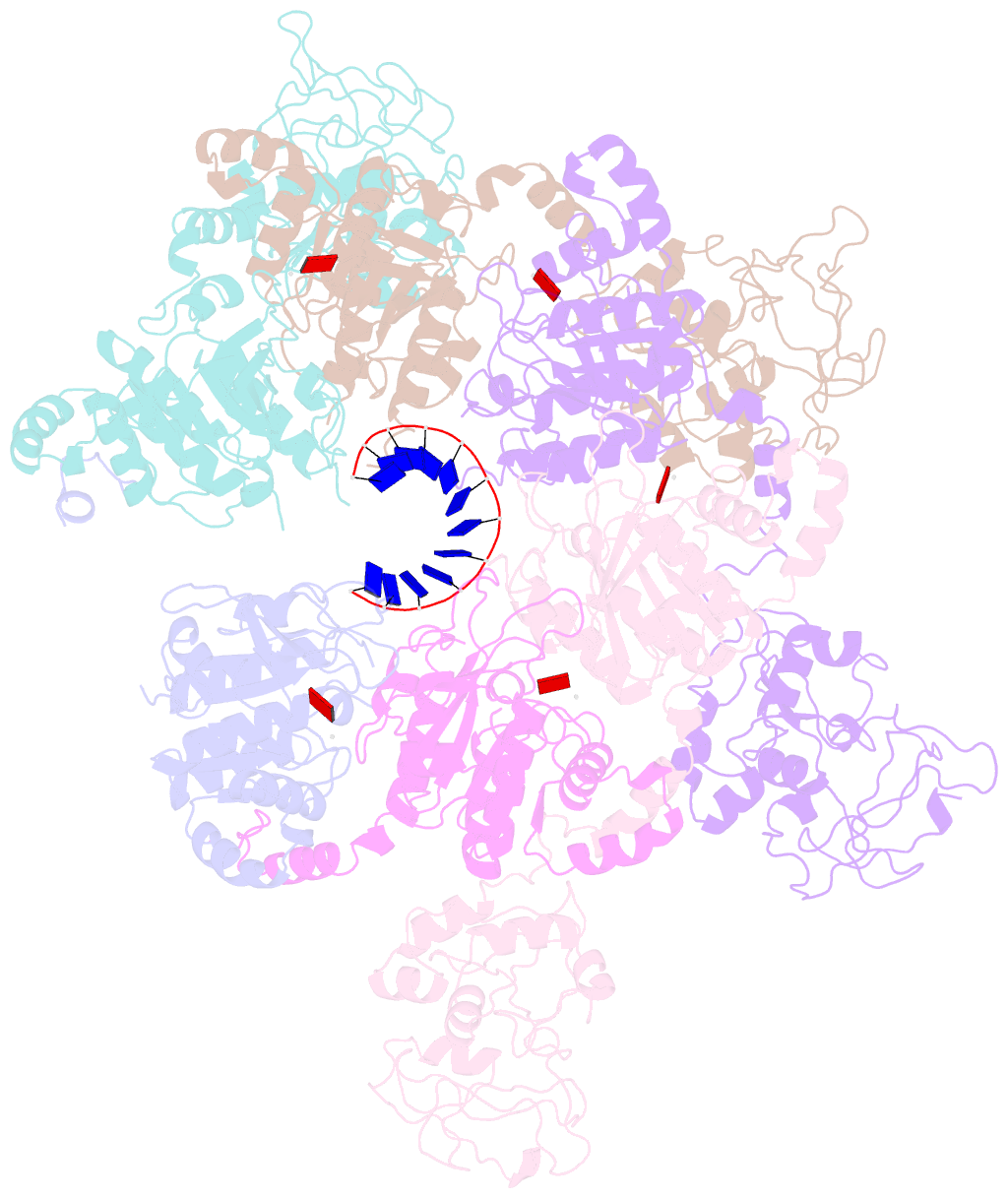
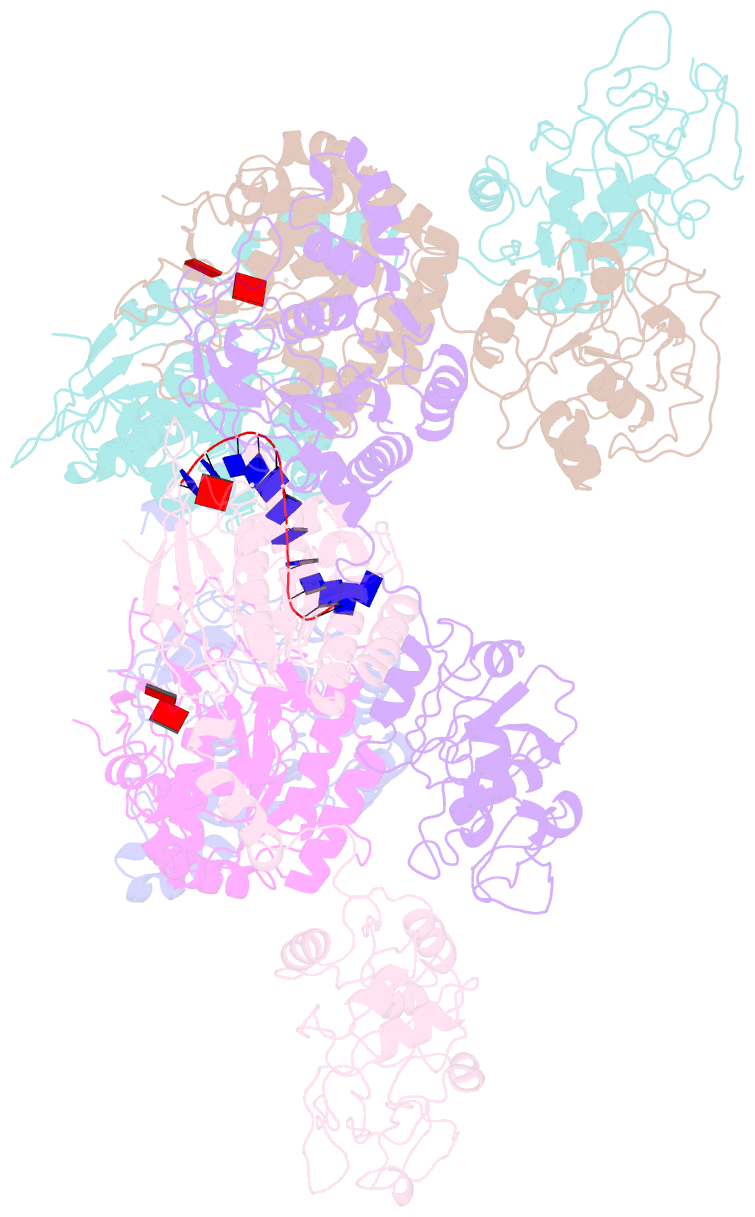
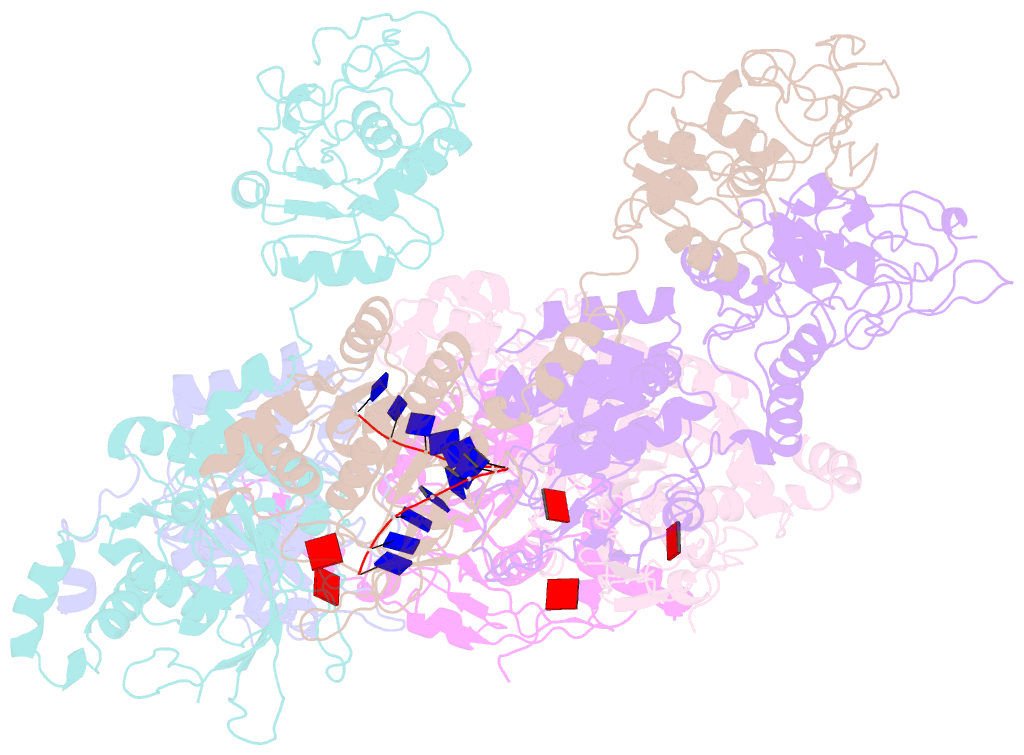
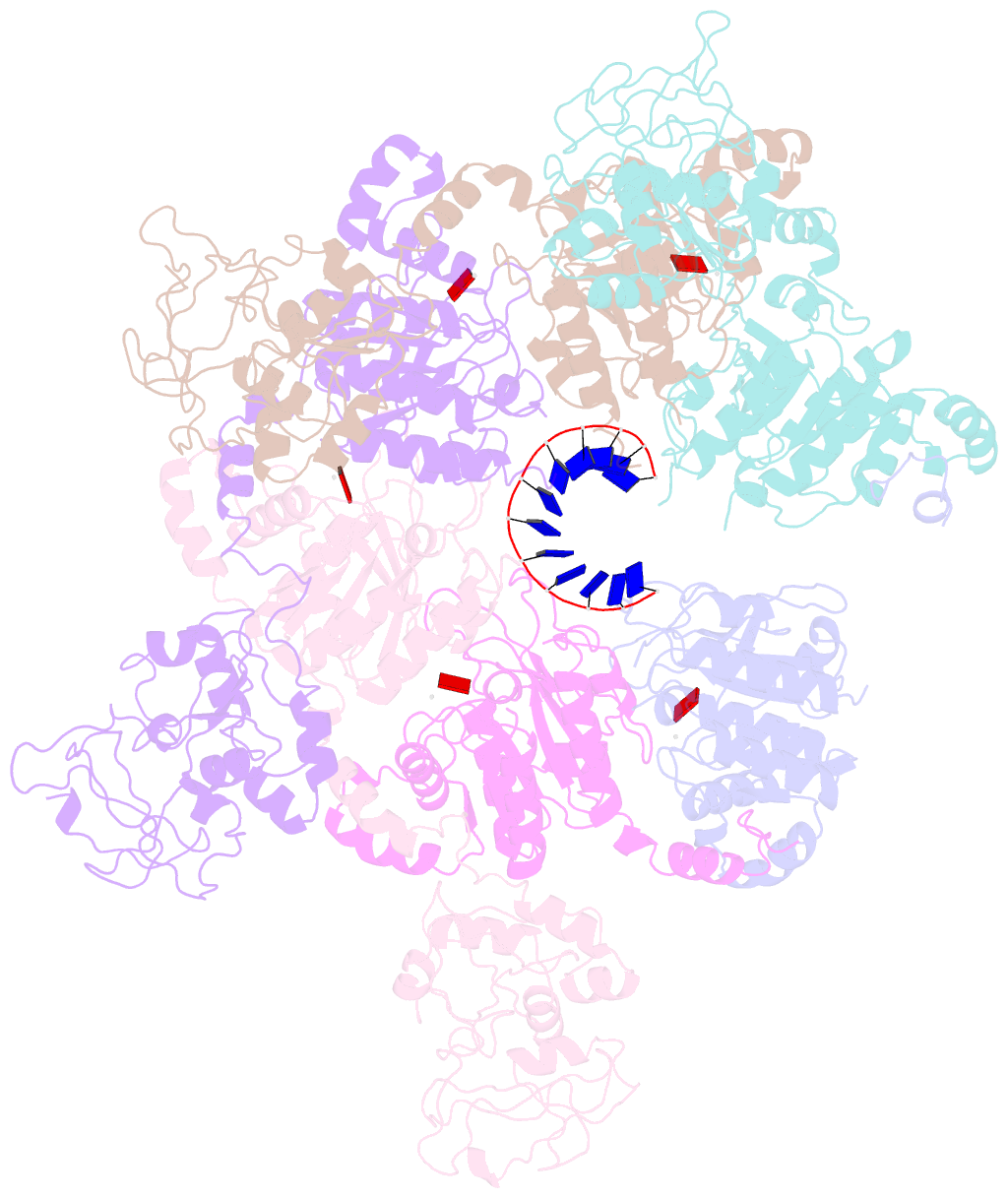
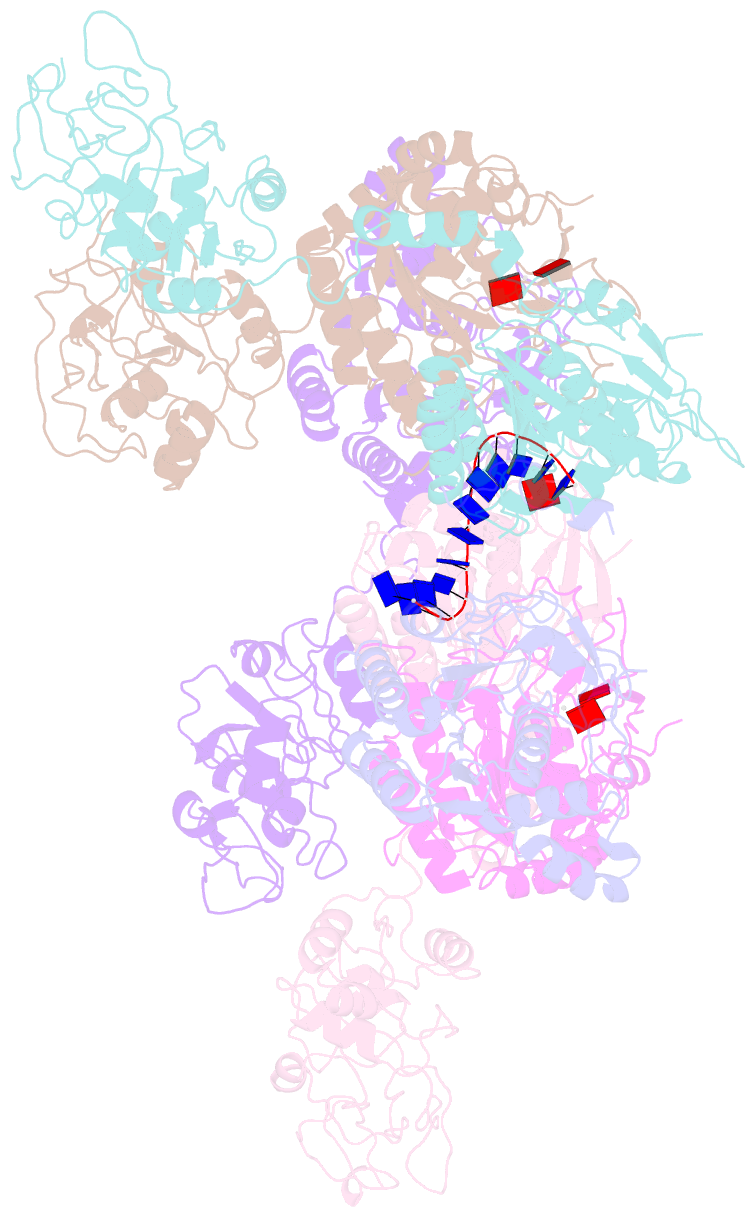
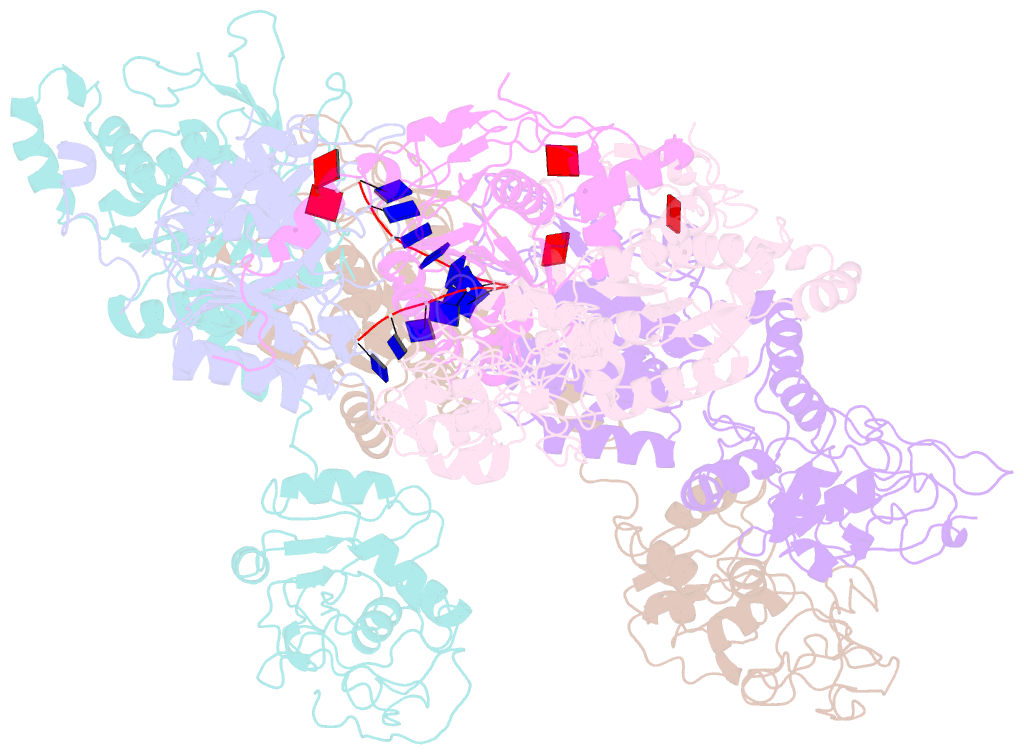
- Structure of lates calcarifer twinkle helicase with atp and DNA
- Reference
- Li Z, Kaur P, Lo CY, Chopra N, Smith J, Wang H, Gao Y (2022): "Structural and dynamic basis of DNA capture and translocation by mitochondrial Twinkle helicase." Nucleic Acids Res., 50, 11965-11978. doi: 10.1093/nar/gkac1089.
- Abstract
- Twinkle is a mitochondrial replicative helicase which can self-load onto and unwind mitochondrial DNA. Nearly 60 mutations on Twinkle have been linked to human mitochondrial diseases. Using cryo-electron microscopy (cryo-EM) and high-speed atomic force microscopy (HS-AFM), we obtained the atomic-resolution structure of a vertebrate Twinkle homolog with DNA and captured in real-time how Twinkle is self-loaded onto DNA. Our data highlight the important role of the non-catalytic N-terminal domain of Twinkle. The N-terminal domain directly contacts the C-terminal helicase domain, and the contact interface is a hotspot for disease-related mutations. Mutations at the interface destabilize Twinkle hexamer and reduce helicase activity. With HS-AFM, we observed that a highly dynamic Twinkle domain, which is likely to be the N-terminal domain, can protrude ∼5 nm to transiently capture nearby DNA and initialize Twinkle loading onto DNA. Moreover, structural analysis and subunit doping experiments suggest that Twinkle hydrolyzes ATP stochastically, which is distinct from related helicases from bacteriophages.