Summary information and primary citation
- PDB-id
- 8e4x; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- X-ray (2.8 Å)
- Summary
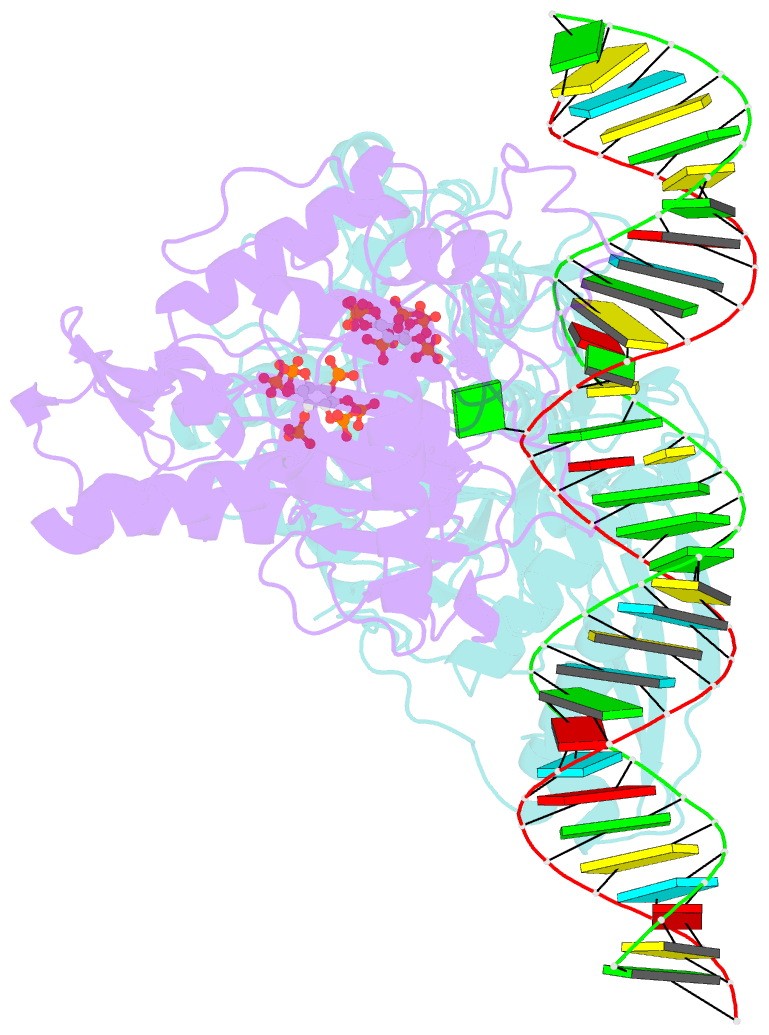
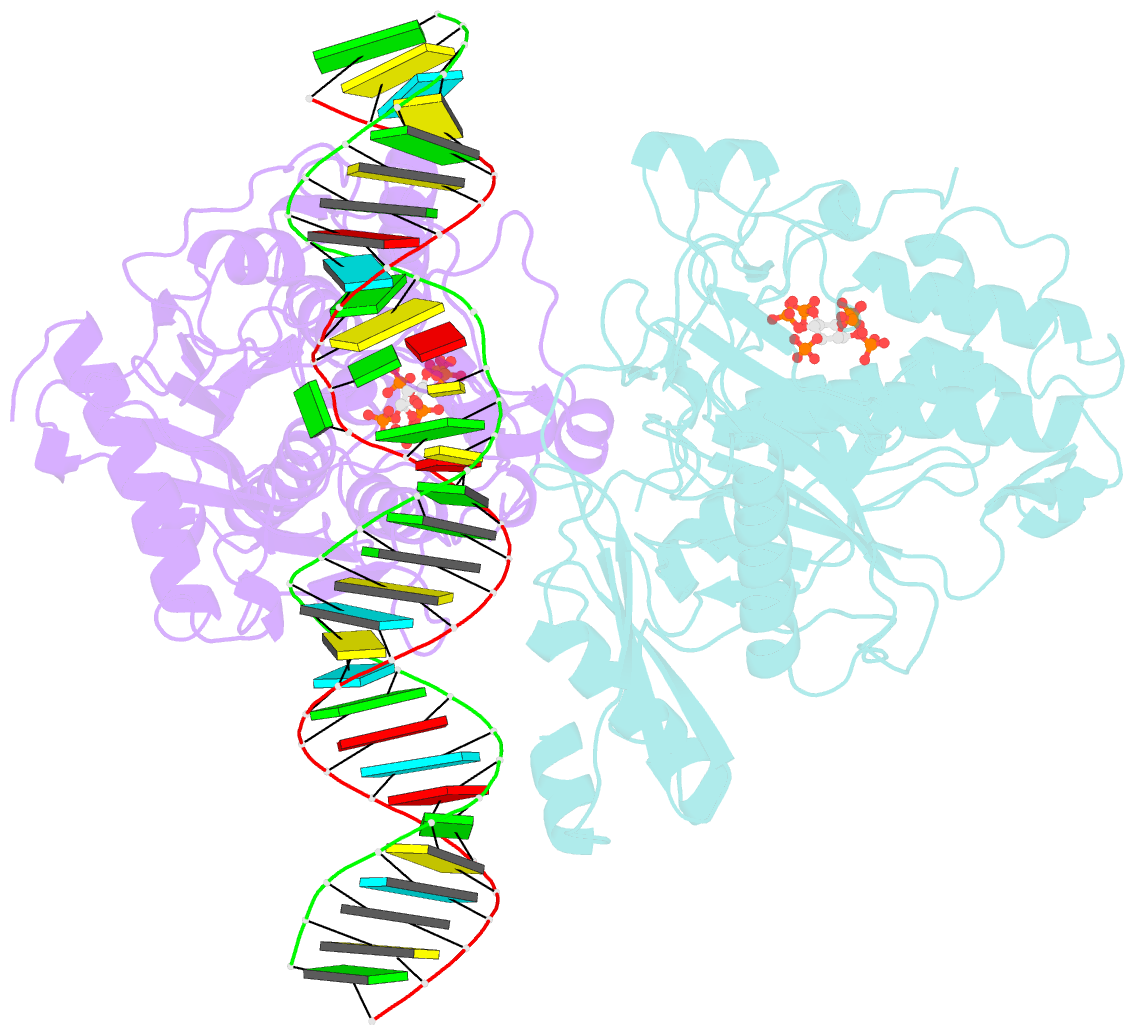
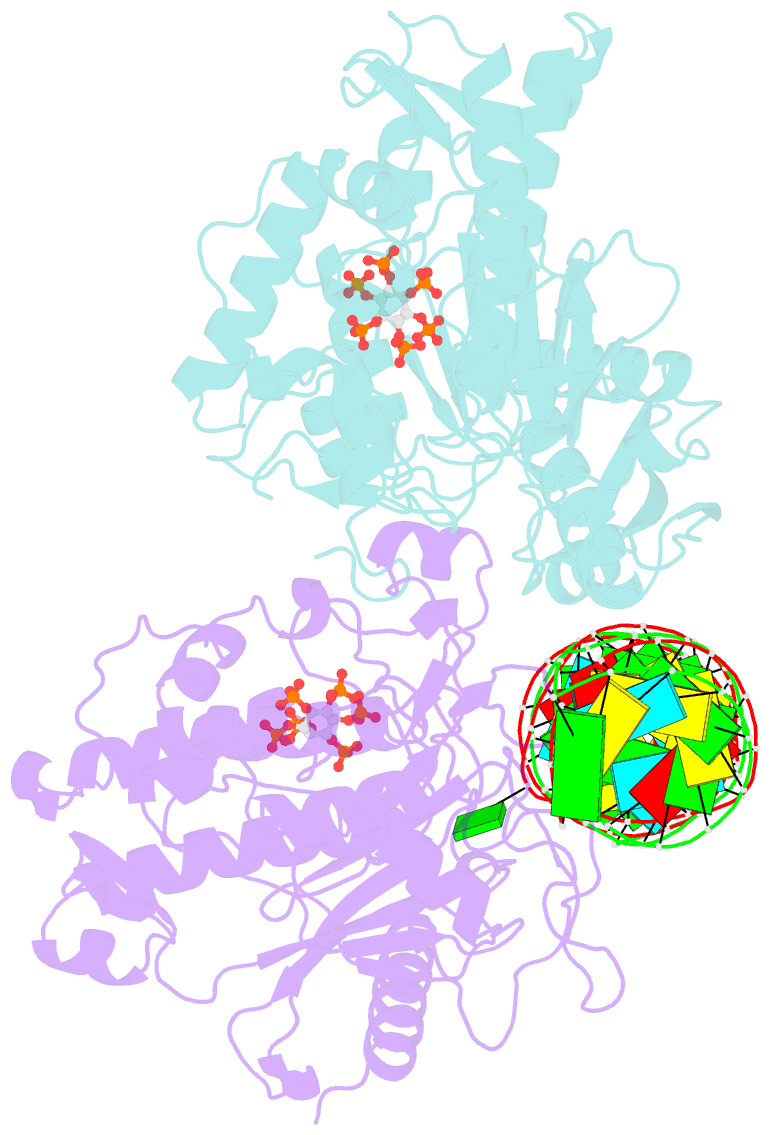
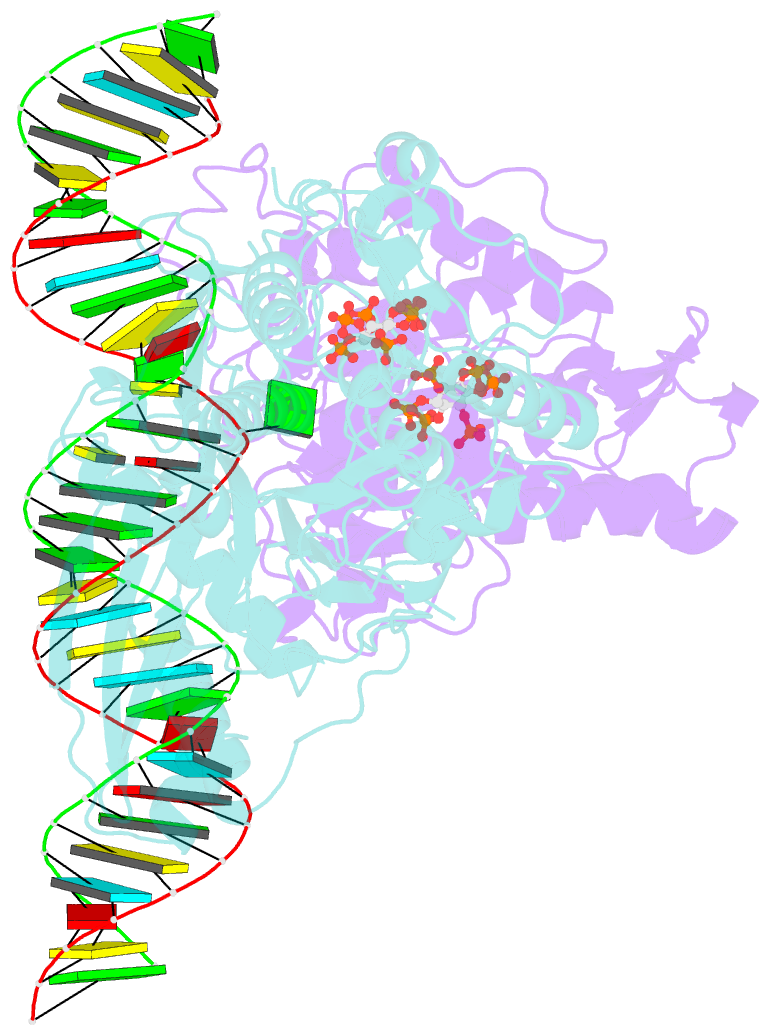
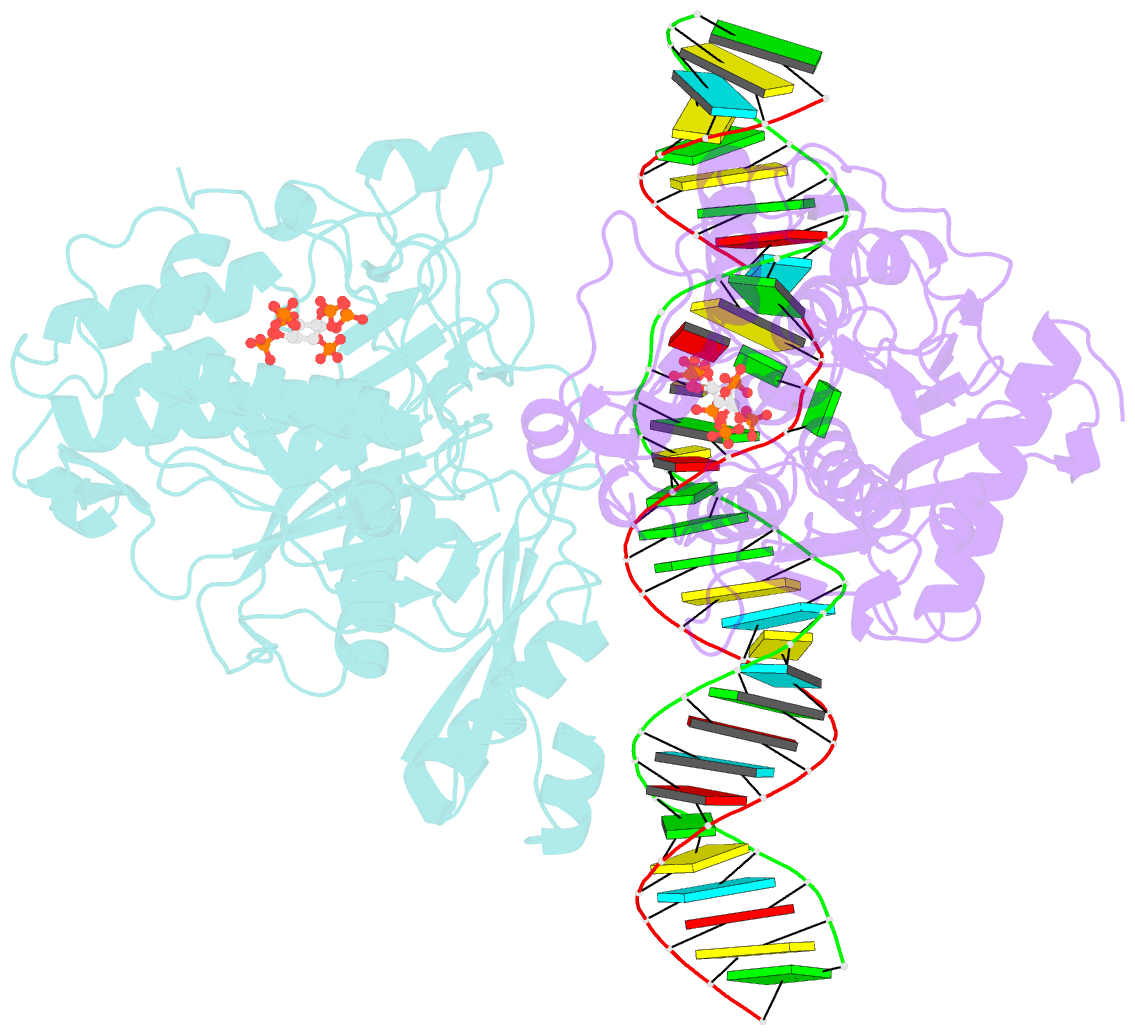
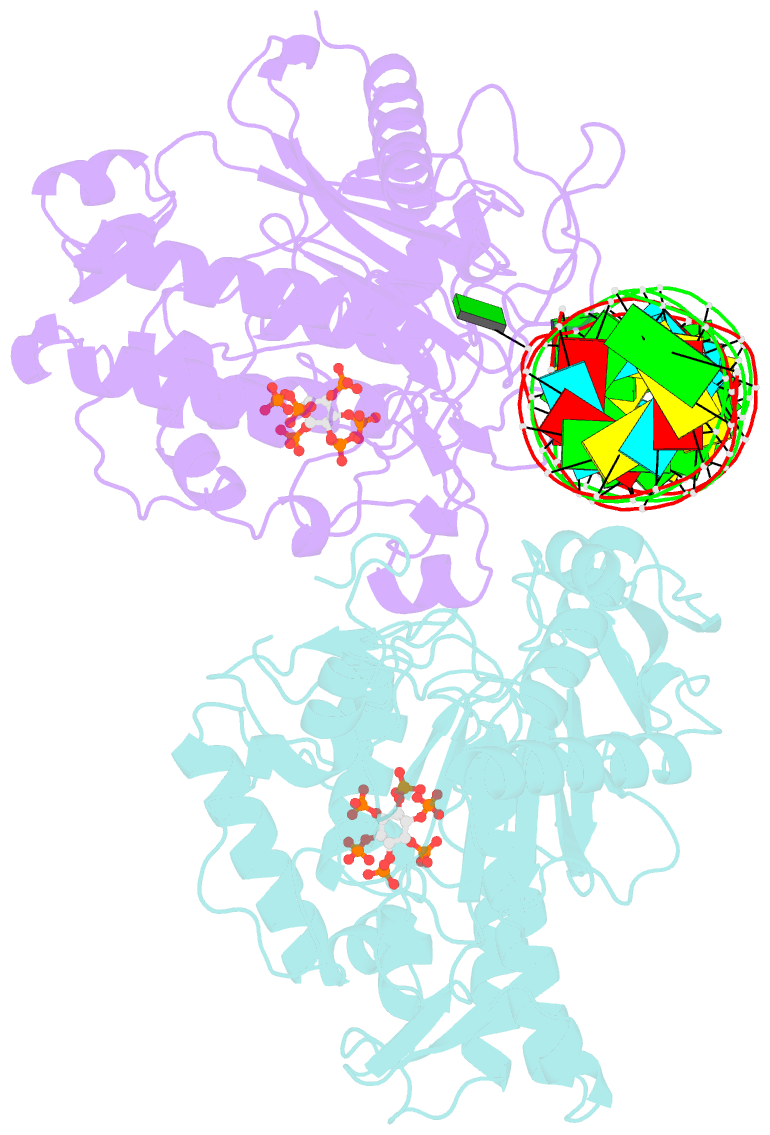
- Human adenosine deaminase acting on dsrna (adar2-r2d) bound to dsrna containing a g:3-deaza da pair adjacent to the target site
- Reference
- Doherty EE, Karki A, Wilcox XE, Mendoza HG, Manjunath A, Matos VJ, Fisher AJ, Beal PA (2022): "ADAR activation by inducing a syn conformation at guanosine adjacent to an editing site." Nucleic Acids Res., 50, 10857-10868. doi: 10.1093/nar/gkac897.
- Abstract
- ADARs (adenosine deaminases acting on RNA) can be directed to sites in the transcriptome by complementary guide strands allowing for the correction of disease-causing mutations at the RNA level. However, ADARs show bias against editing adenosines with a guanosine 5' nearest neighbor (5'-GA sites), limiting the scope of this approach. Earlier studies suggested this effect arises from a clash in the RNA minor groove involving the 2-amino group of the guanosine adjacent to an editing site. Here we show that nucleosides capable of pairing with guanosine in a syn conformation enhance editing for 5'-GA sites. We describe the crystal structure of a fragment of human ADAR2 bound to RNA bearing a G:G pair adjacent to an editing site. The two guanosines form a Gsyn:Ganti pair solving the steric problem by flipping the 2-amino group of the guanosine adjacent to the editing site into the major groove. Also, duplexes with 2'-deoxyadenosine and 3-deaza-2'-deoxyadenosine displayed increased editing efficiency, suggesting the formation of a Gsyn:AH+anti pair. This was supported by X-ray crystallography of an ADAR complex with RNA bearing a G:3-deaza dA pair. This study shows how non-Watson-Crick pairing in duplex RNA can facilitate ADAR editing enabling the design of next generation guide strands for therapeutic RNA editing.