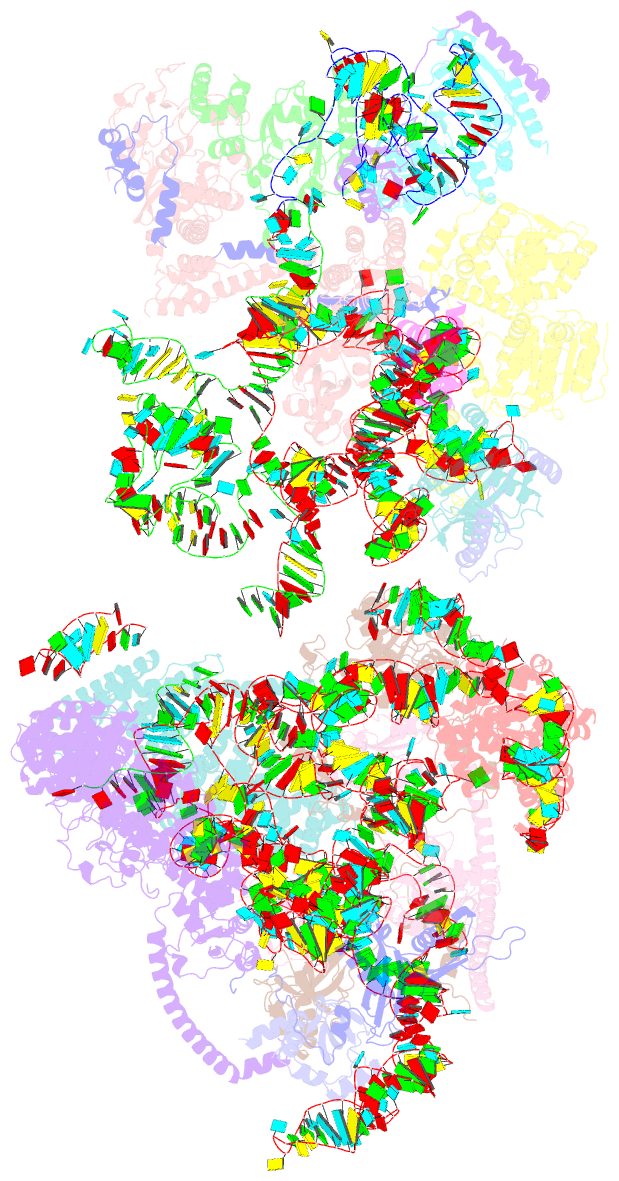
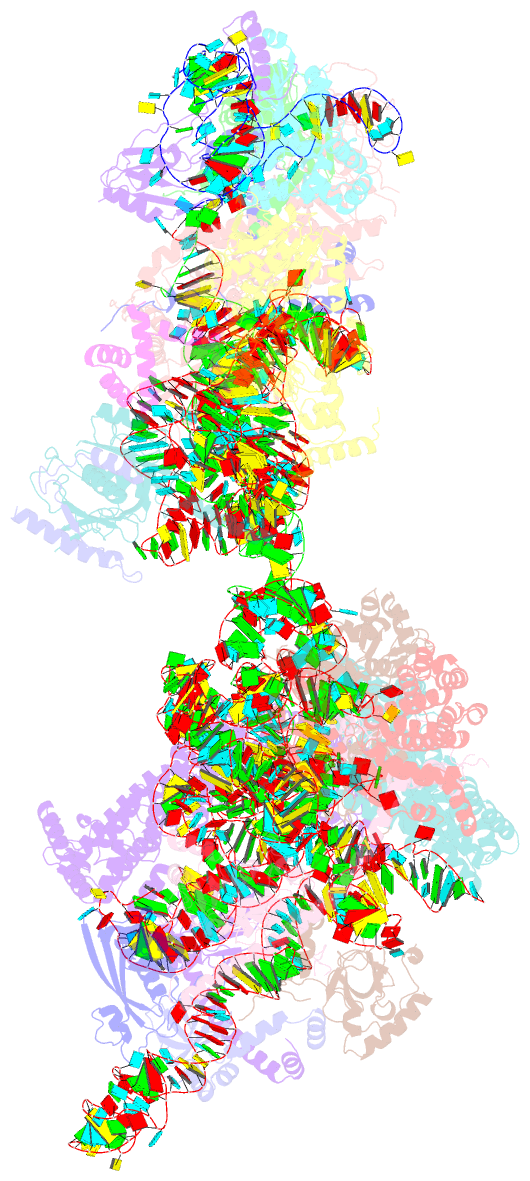
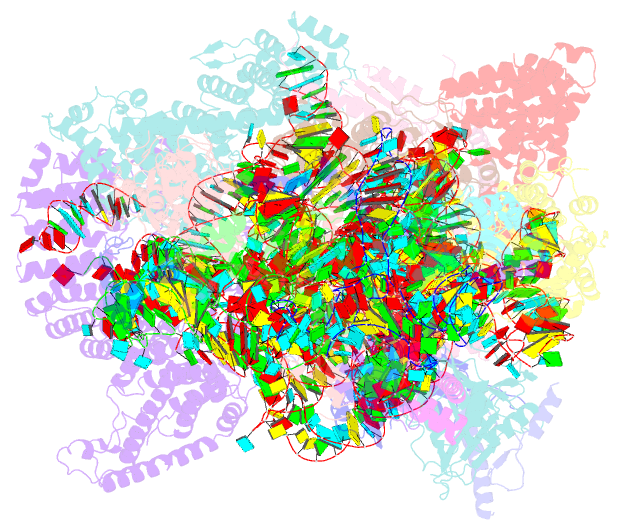
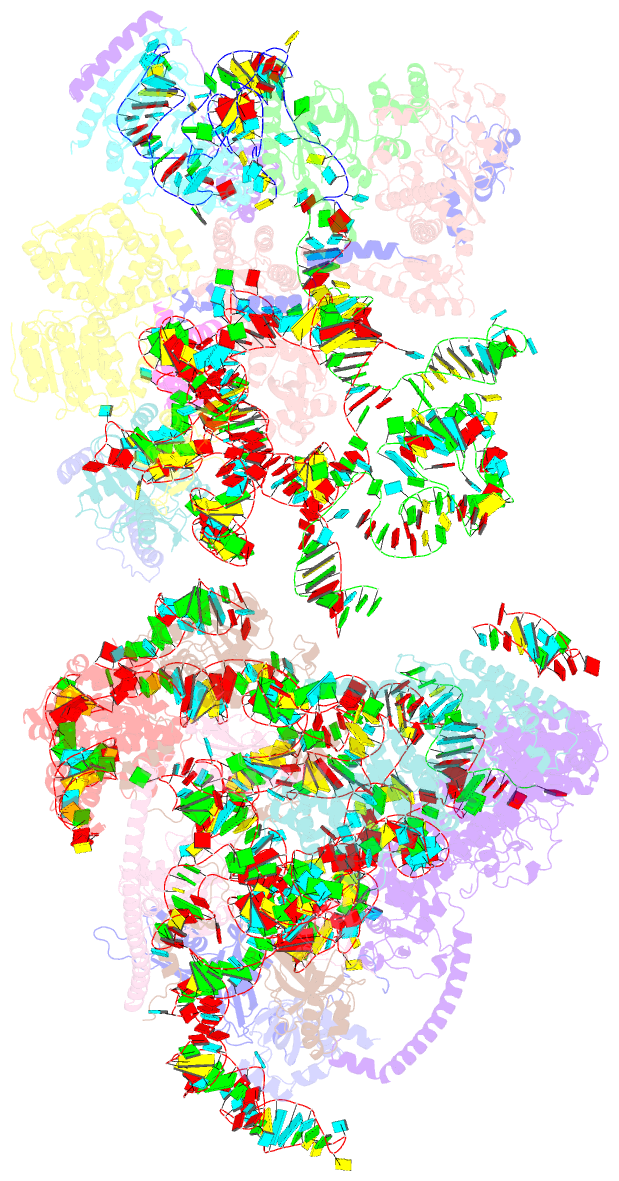
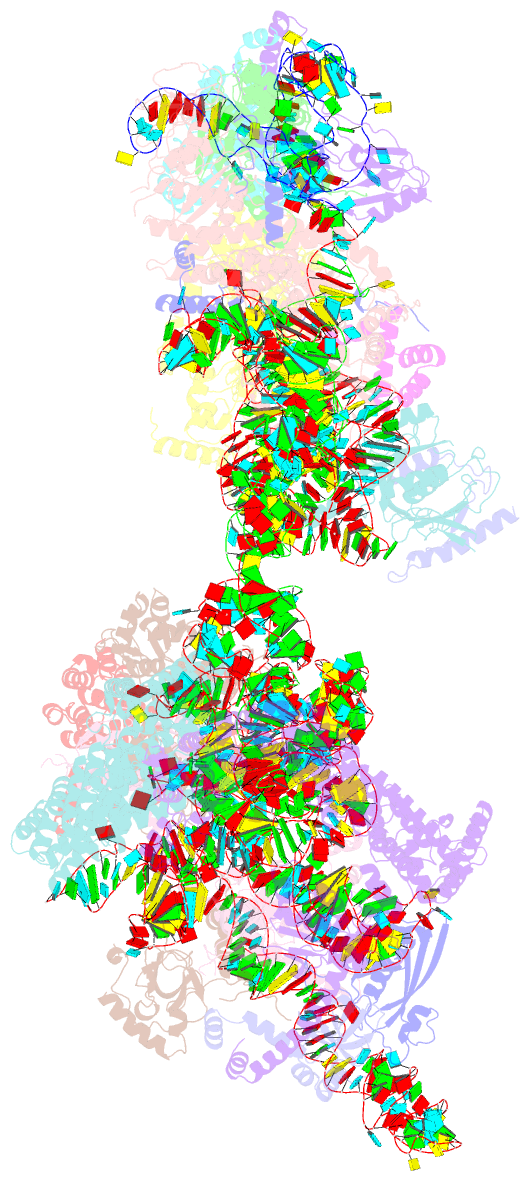
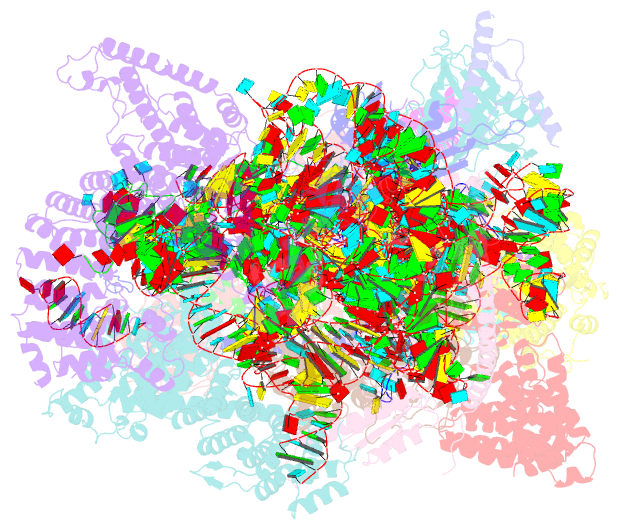
Summary information and primary citation
- PDB-id
- 8e5t; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ribosome
- Method
- cryo-EM (4.0 Å)
- Summary
- Yeast co-transcriptional noc1-noc2 rnp assembly checkpoint intermediate
- Reference
- Sanghai ZA, Piwowarczyk R, Broeck AV, Klinge S (2023): "A co-transcriptional ribosome assembly checkpoint controls nascent large ribosomal subunit maturation." Nat.Struct.Mol.Biol., 30, 594-599. doi: 10.1038/s41594-023-00947-3.
- Abstract
- During transcription of eukaryotic ribosomal DNA in the nucleolus, assembly checkpoints exist that guarantee the formation of stable precursors of small and large ribosomal subunits. While the formation of an early large subunit assembly checkpoint precedes the separation of small and large subunit maturation, its mechanism of action and function remain unknown. Here, we report the cryo-electron microscopy structure of the yeast co-transcriptional large ribosomal subunit assembly intermediate that serves as a checkpoint. The structure provides the mechanistic basis for how quality-control pathways are established through co-transcriptional ribosome assembly factors, that structurally interrogate, remodel and, together with ribosomal proteins, cooperatively stabilize correctly folded pre-ribosomal RNA. Our findings thus provide a molecular explanation for quality control during eukaryotic ribosome assembly in the nucleolus.