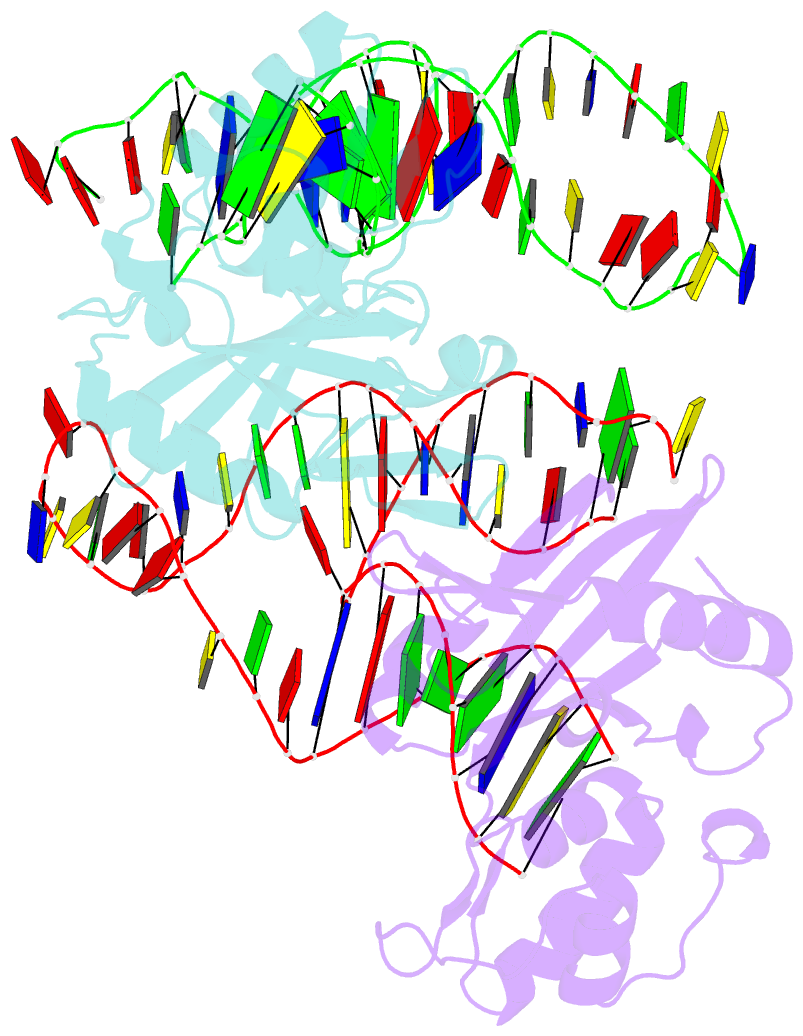
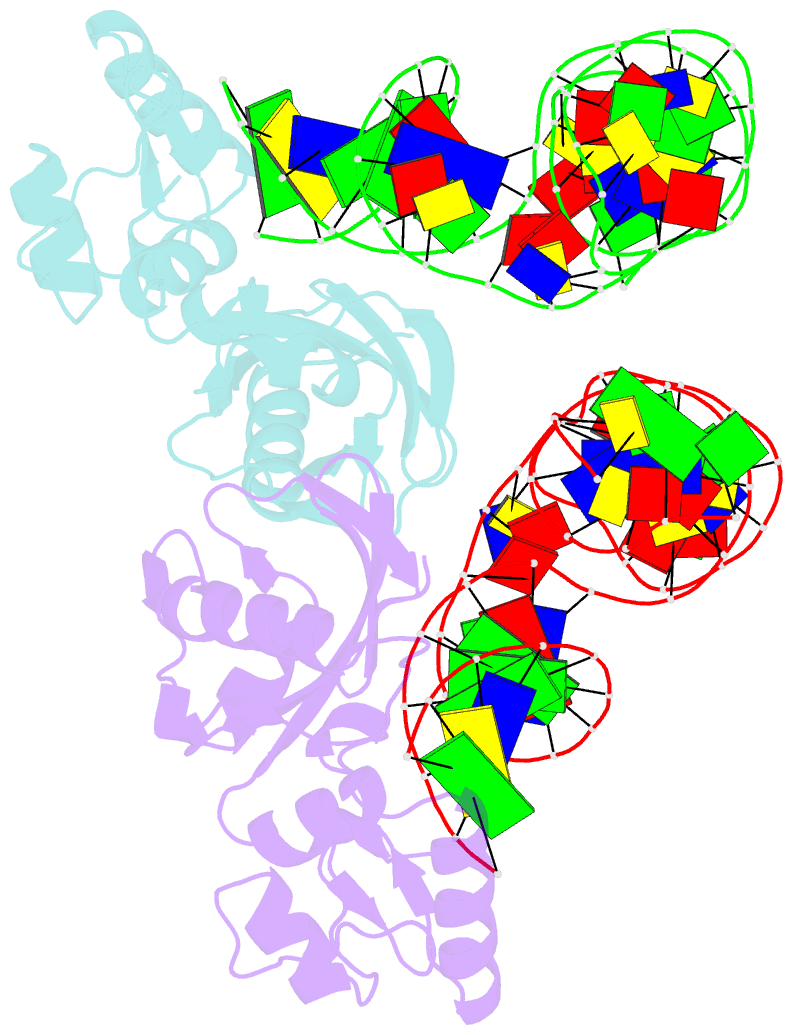
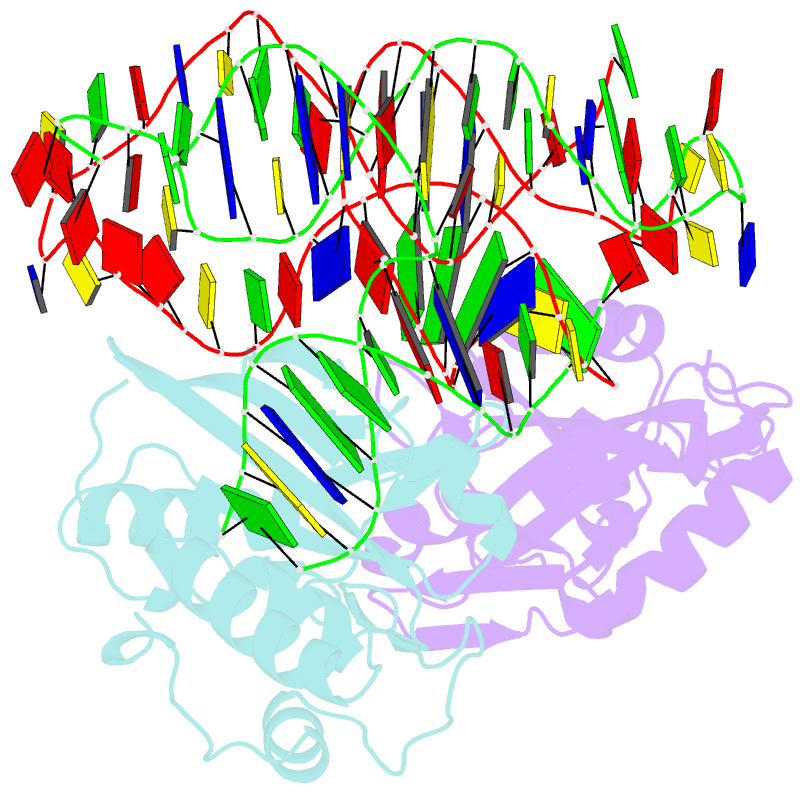
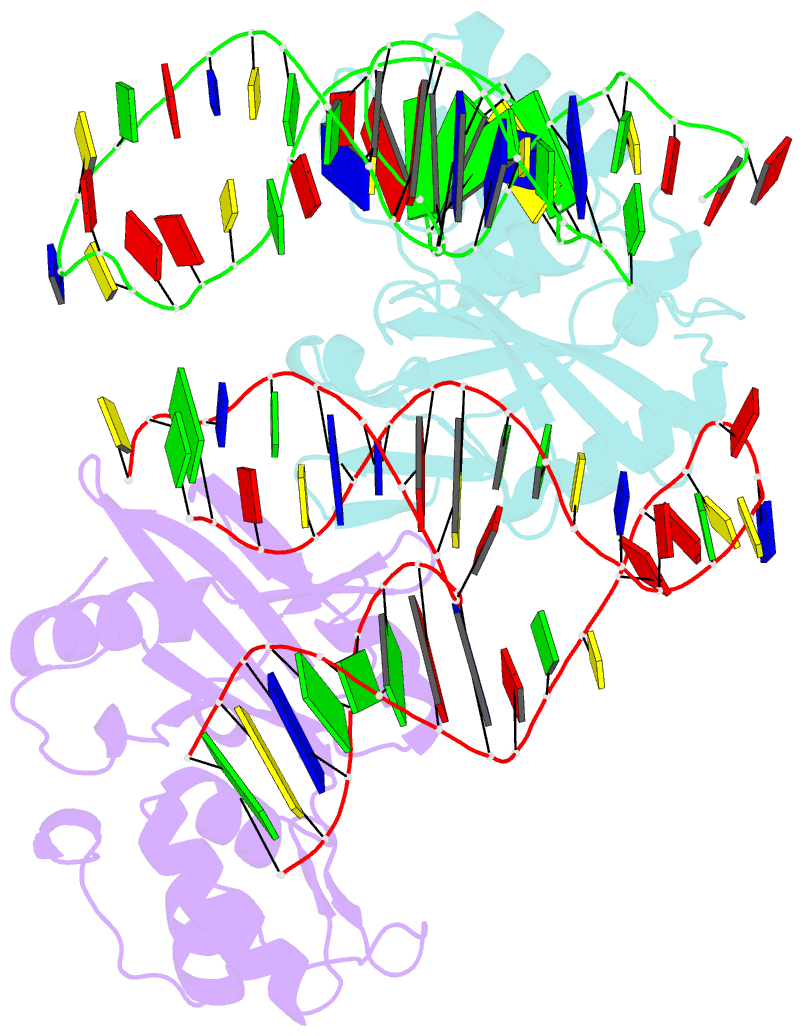
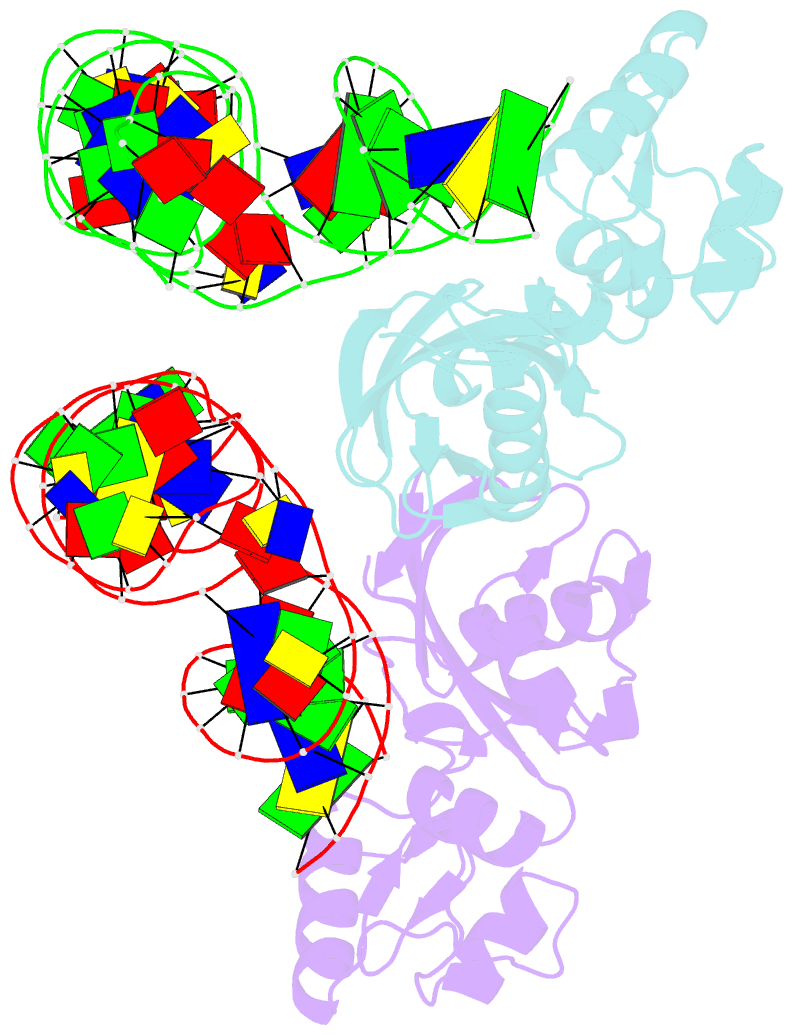
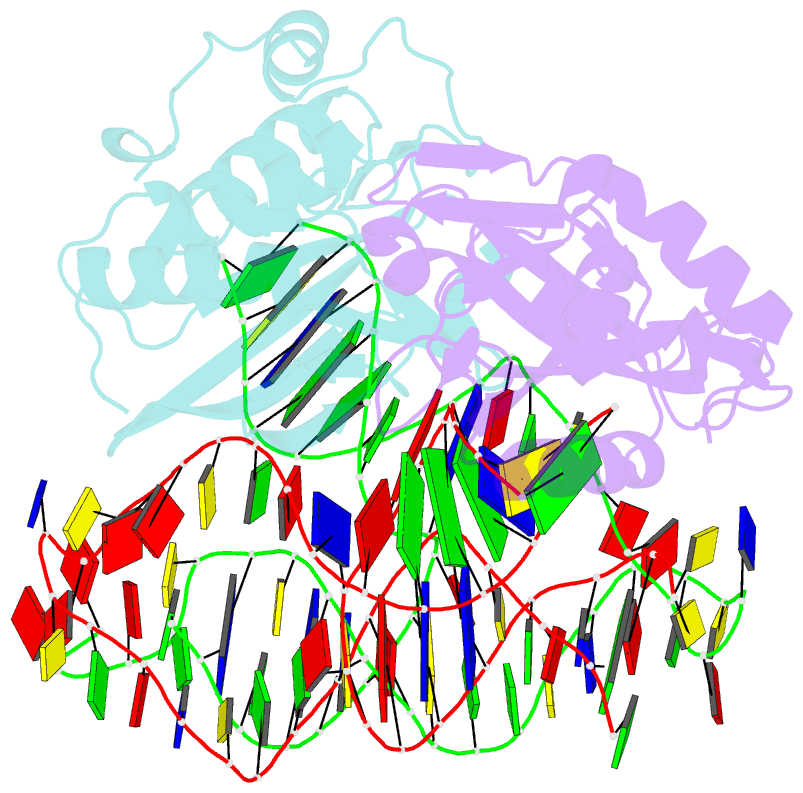
Summary information and primary citation
- PDB-id
- 8e9a; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.69 Å)
- Summary
- Crystal structure of asfvpolx in complex with 10-23 dnazyme and mg
- Reference
- Cramer ER, Starcovic SA, Avey RM, Kaya AI, Robart AR (2023): "Structure of a 10-23 deoxyribozyme exhibiting a homodimer conformation." Commun Chem, 6, 119. doi: 10.1038/s42004-023-00924-3.
- Abstract
- Deoxyribozymes (DNAzymes) are in vitro evolved DNA sequences capable of catalyzing chemical reactions. The RNA-cleaving 10-23 DNAzyme was the first DNAzyme to be evolved and possesses clinical and biotechnical applications as a biosensor and a knockdown agent. DNAzymes do not require the recruitment of other components to cleave RNA and can turnover, thus they have a distinct advantage over other knockdown methods (siRNA, CRISPR, morpholinos). Despite this, a lack of structural and mechanistic information has hindered the optimization and application of the 10-23 DNAzyme. Here, we report a 2.7 Å crystal structure of the RNA-cleaving 10-23 DNAzyme in a homodimer conformation. Although proper coordination of the DNAzyme to substrate is observed along with intriguing patterns of bound magnesium ions, the dimer conformation likely does not capture the true catalytic form of the 10-23 DNAzyme.