Summary information and primary citation
- PDB-id
- 8ebu; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- cryo-EM (3.3 Å)
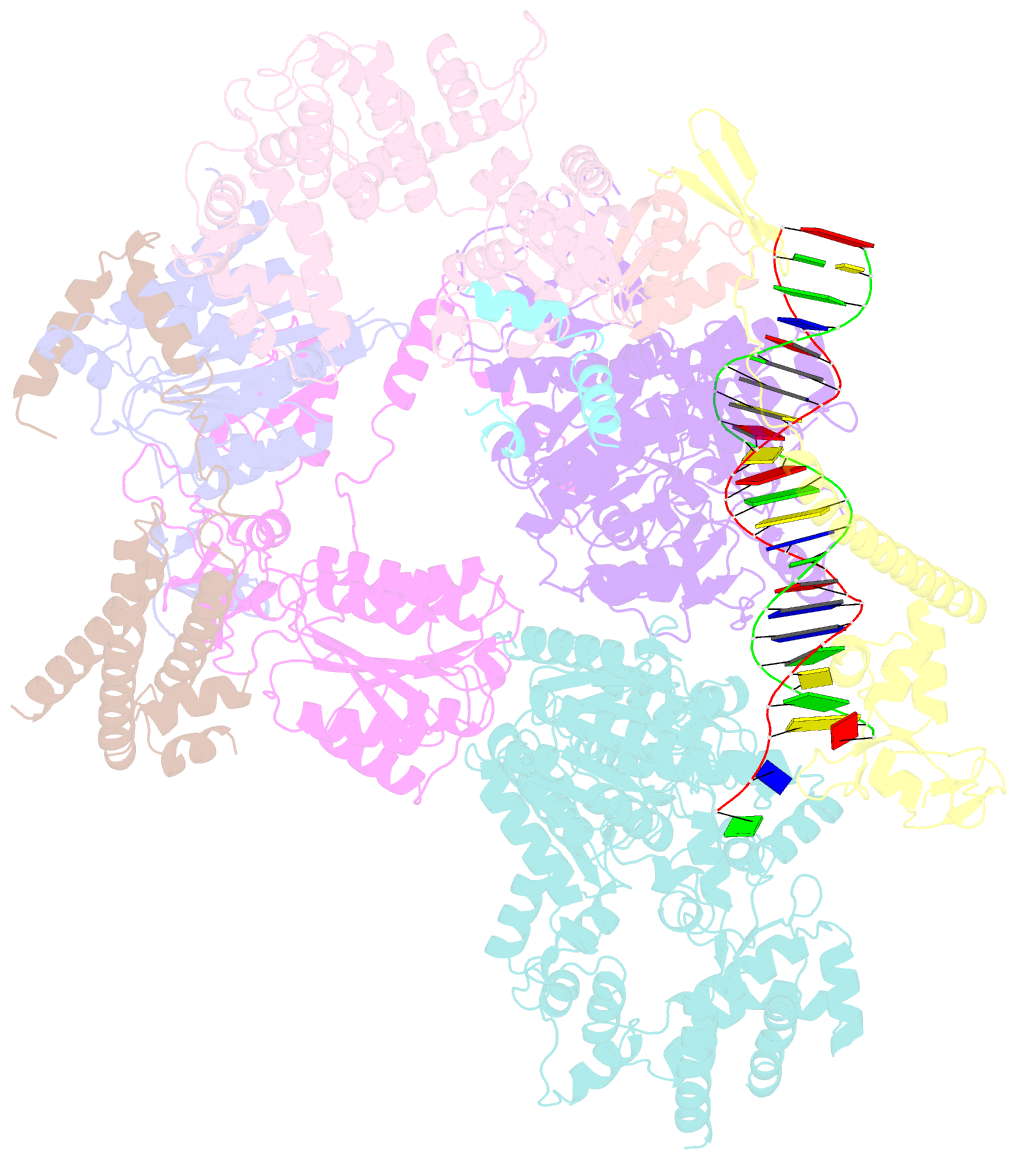
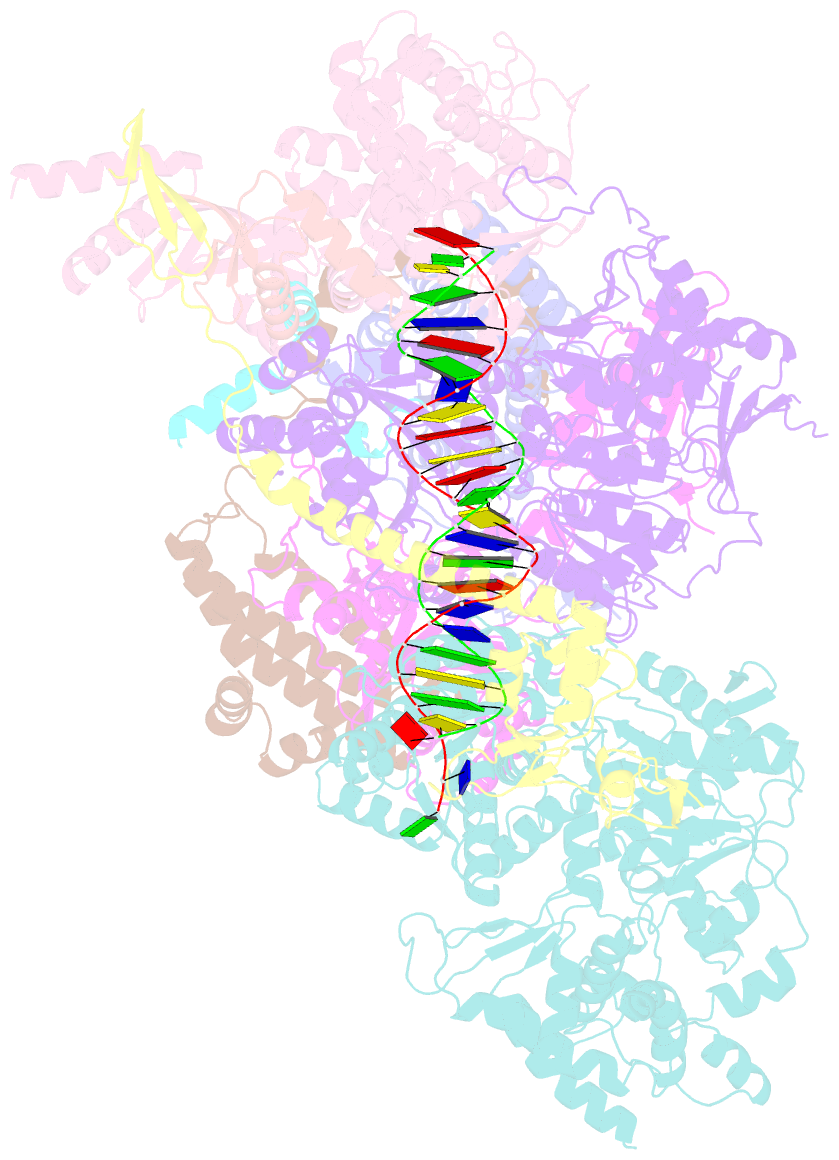
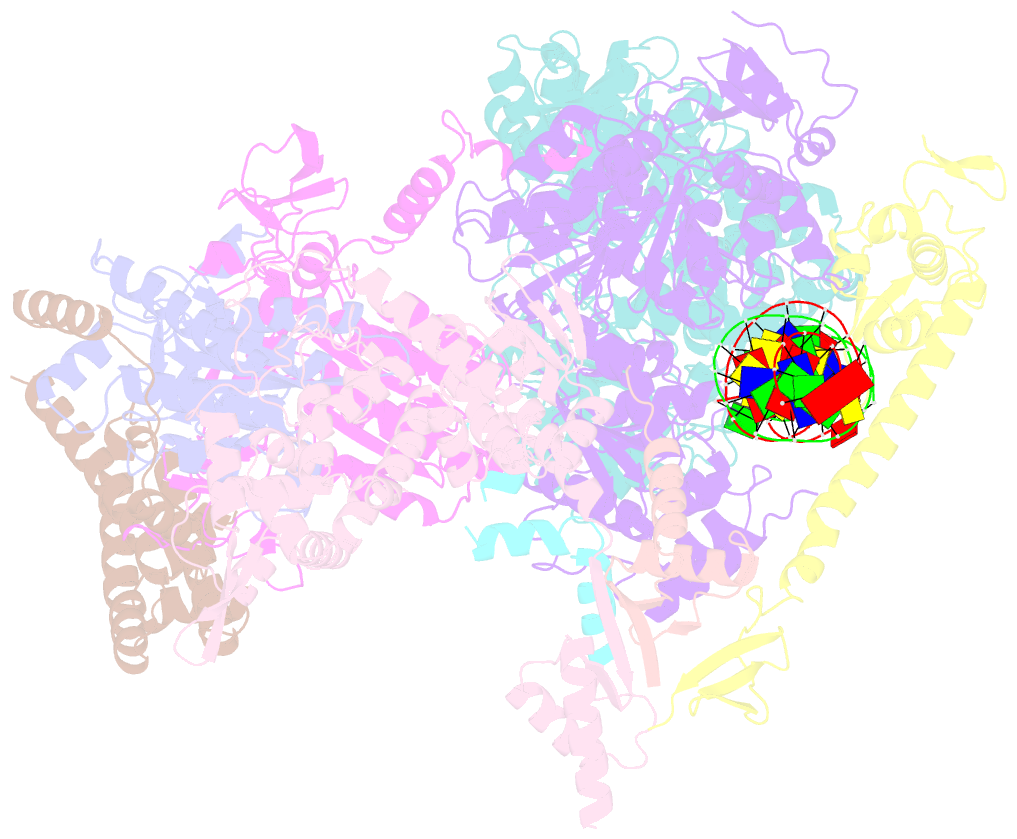
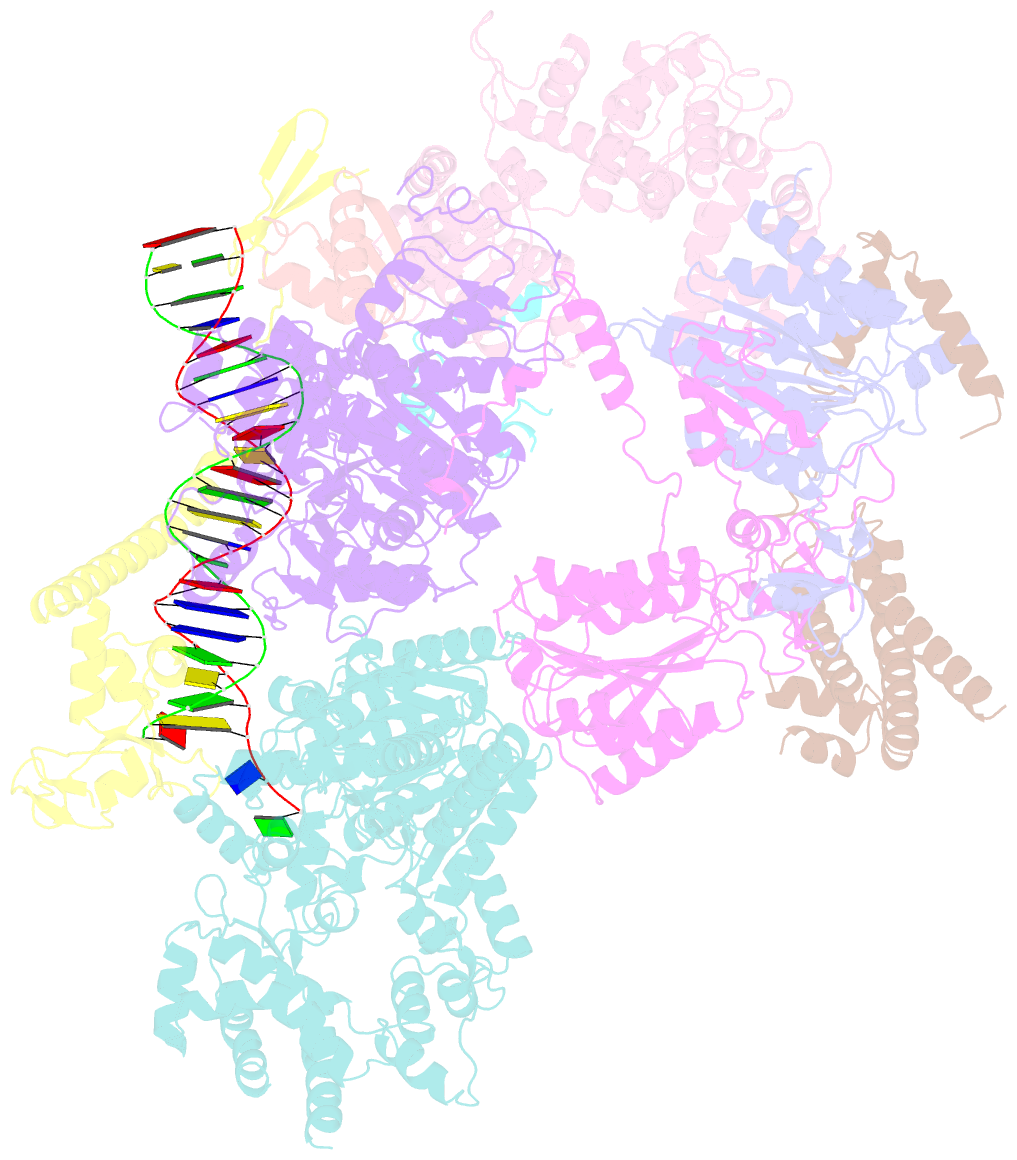
- Summary
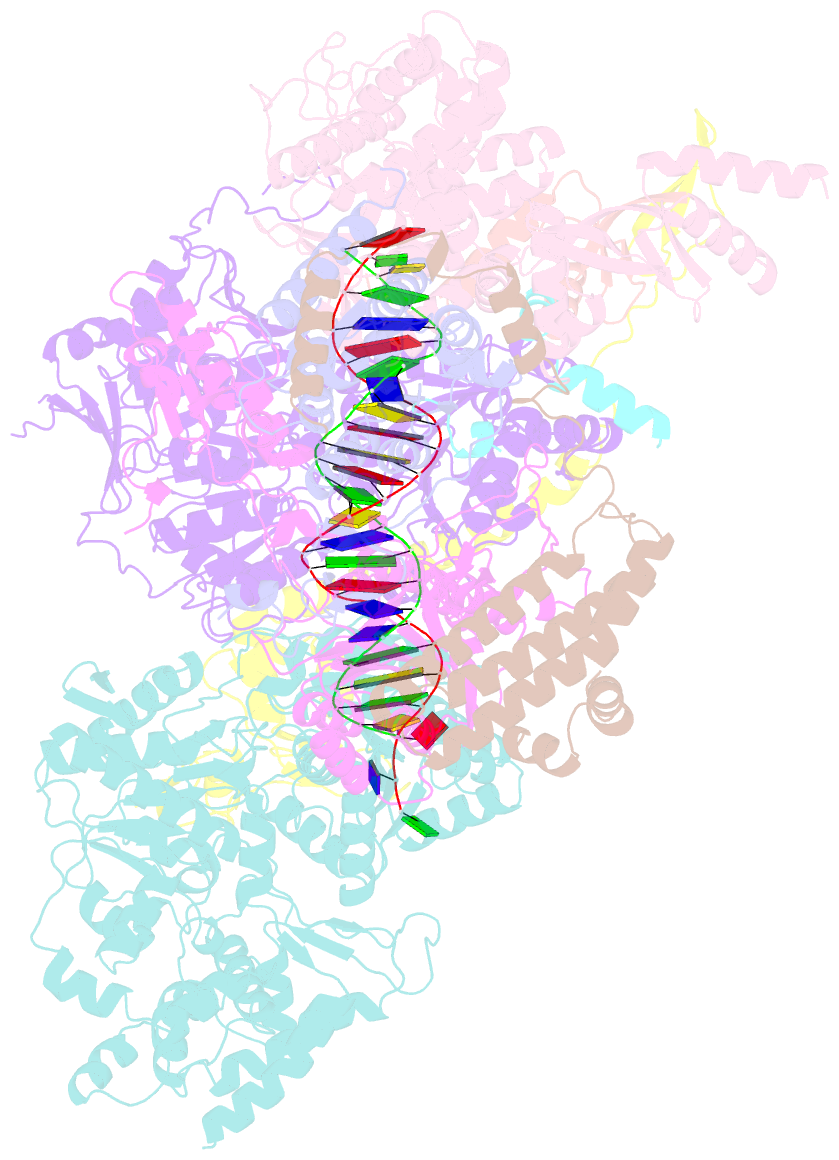
- Xpc release from core7-xpa-DNA (cy5)
- Reference
- Kim J, Li CL, Chen X, Cui Y, Golebiowski FM, Wang H, Hanaoka F, Sugasawa K, Yang W (2023): "Lesion recognition by XPC, TFIIH and XPA in DNA excision repair." Nature, 617, 170-175. doi: 10.1038/s41586-023-05959-z.
- Abstract
- Nucleotide excision repair removes DNA lesions caused by ultraviolet light, cisplatin-like compounds and bulky adducts1. After initial recognition by XPC in global genome repair or a stalled RNA polymerase in transcription-coupled repair, damaged DNA is transferred to the seven-subunit TFIIH core complex (Core7) for verification and dual incisions by the XPF and XPG nucleases2. Structures capturing lesion recognition by the yeast XPC homologue Rad4 and TFIIH in transcription initiation or DNA repair have been separately reported3-7. How two different lesion recognition pathways converge and how the XPB and XPD helicases of Core7 move the DNA lesion for verification are unclear. Here we report on structures revealing DNA lesion recognition by human XPC and DNA lesion hand-off from XPC to Core7 and XPA. XPA, which binds between XPB and XPD, kinks the DNA duplex and shifts XPC and the DNA lesion by nearly a helical turn relative to Core7. The DNA lesion is thus positioned outside of Core7, as would occur with RNA polymerase. XPB and XPD, which track the lesion-containing strand but translocate DNA in opposite directions, push and pull the lesion-containing strand into XPD for verification.