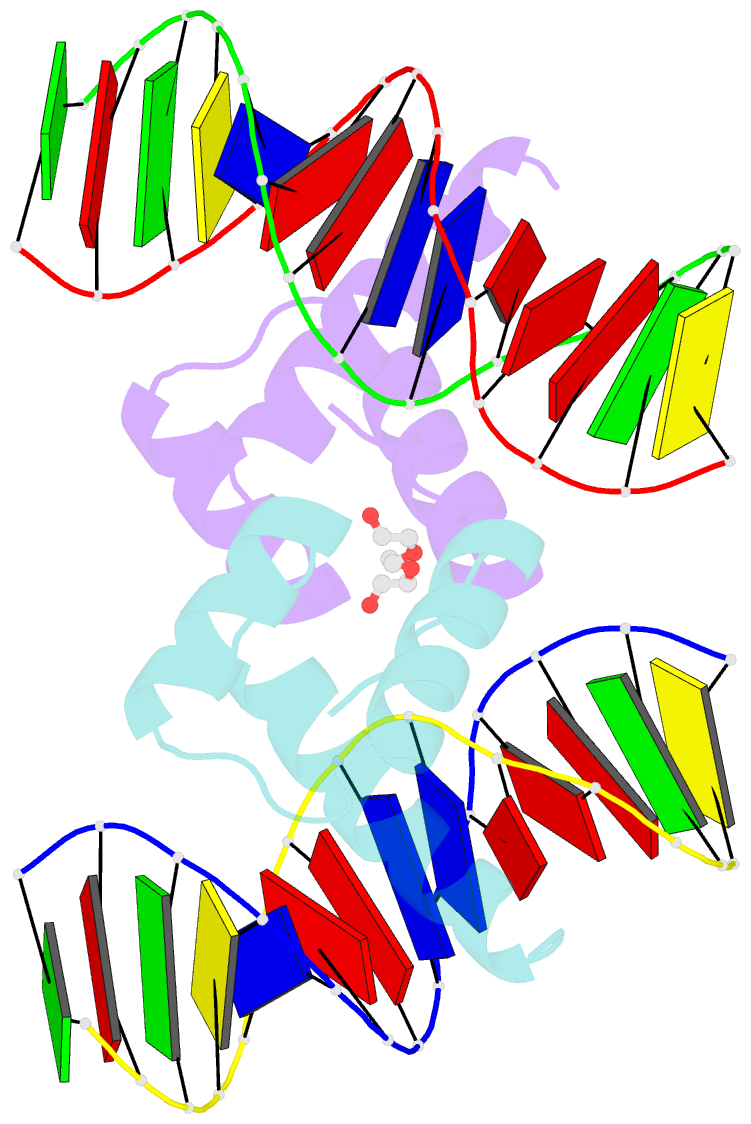
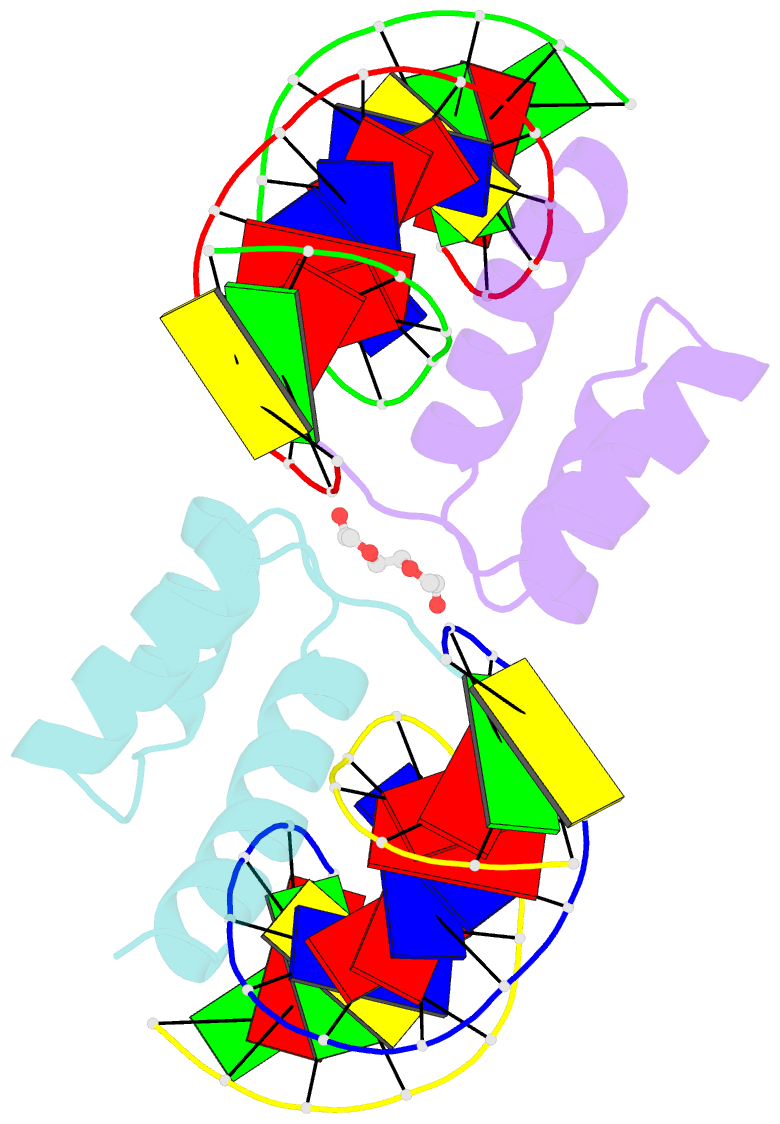
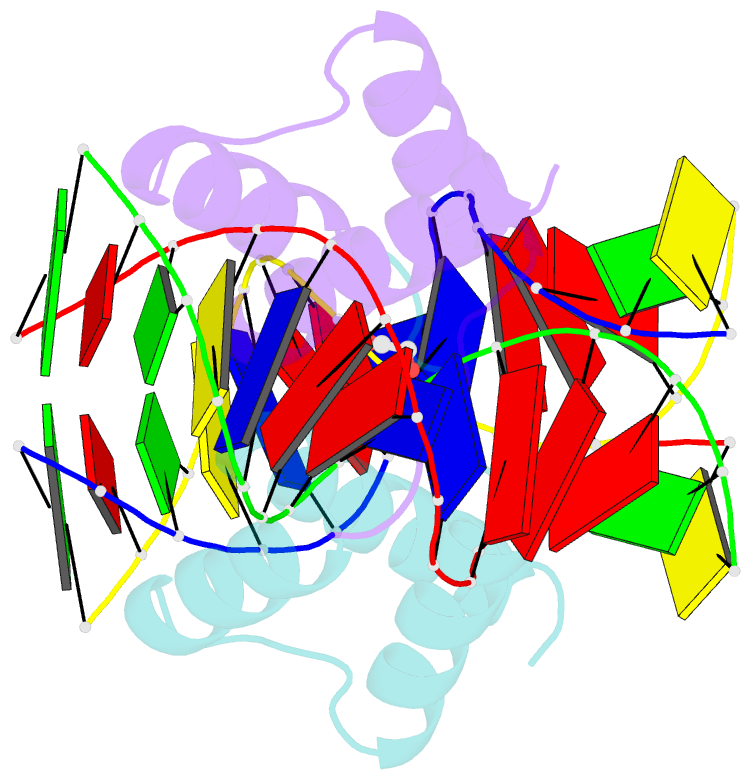
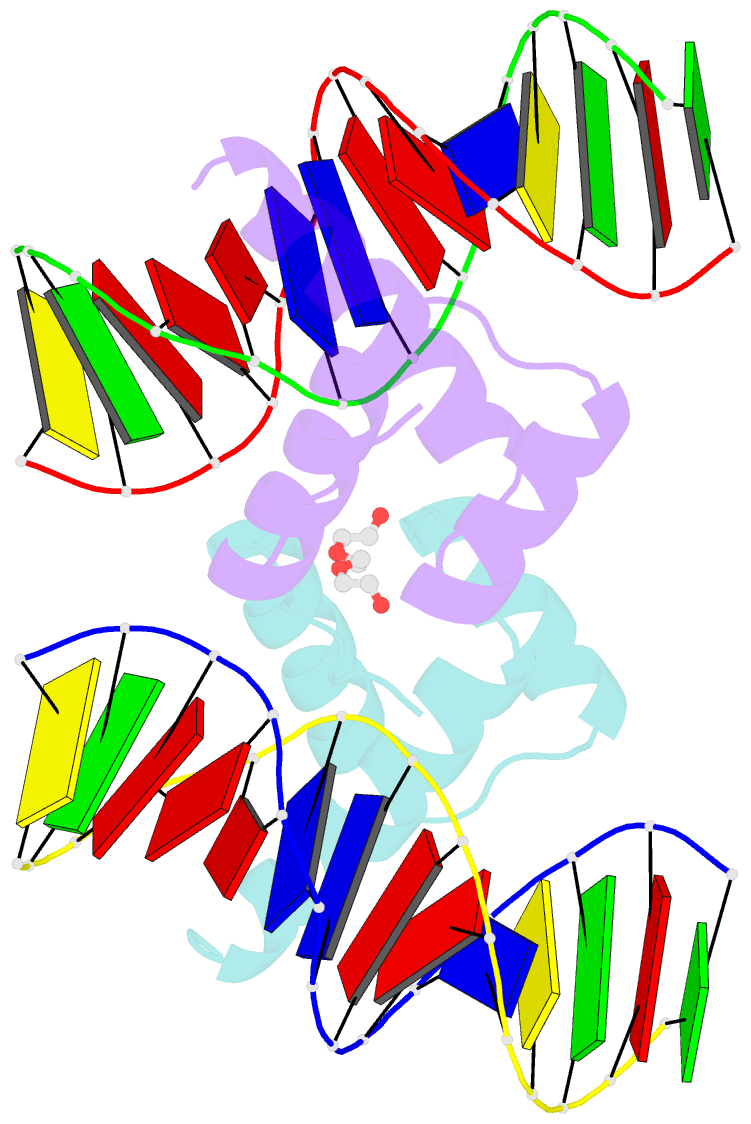
Summary information and primary citation
- PDB-id
- 8eml; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (2.21 Å)
- Summary
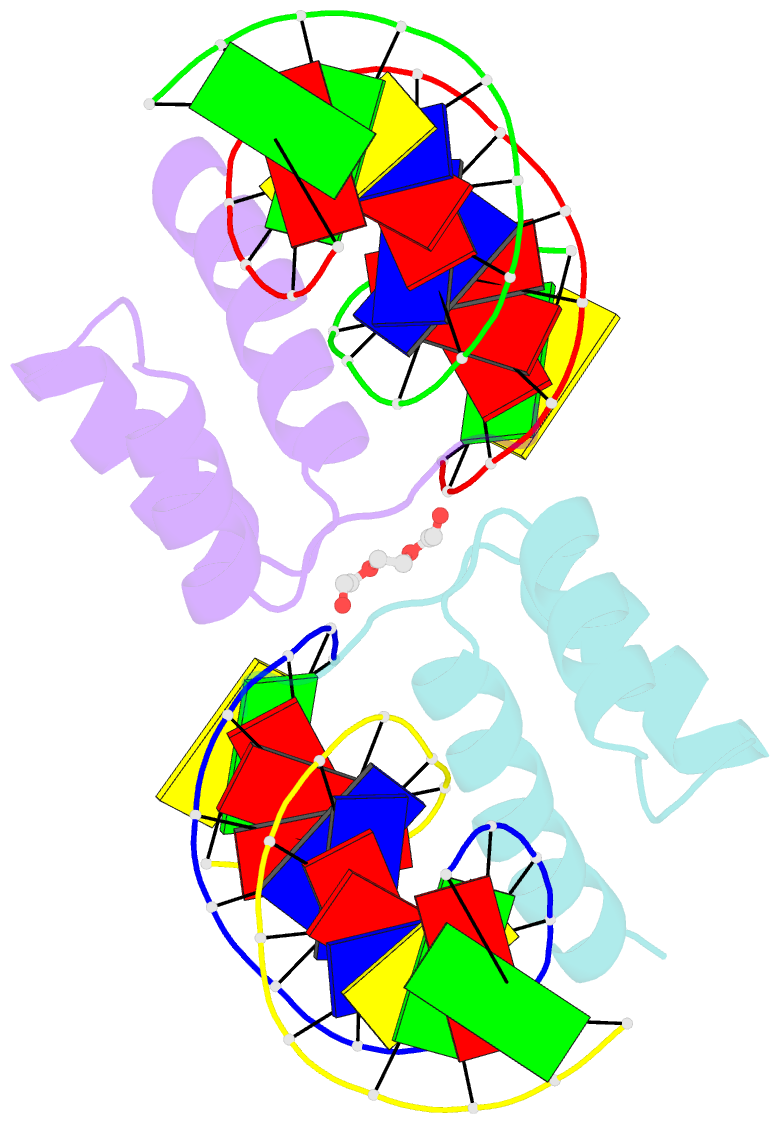
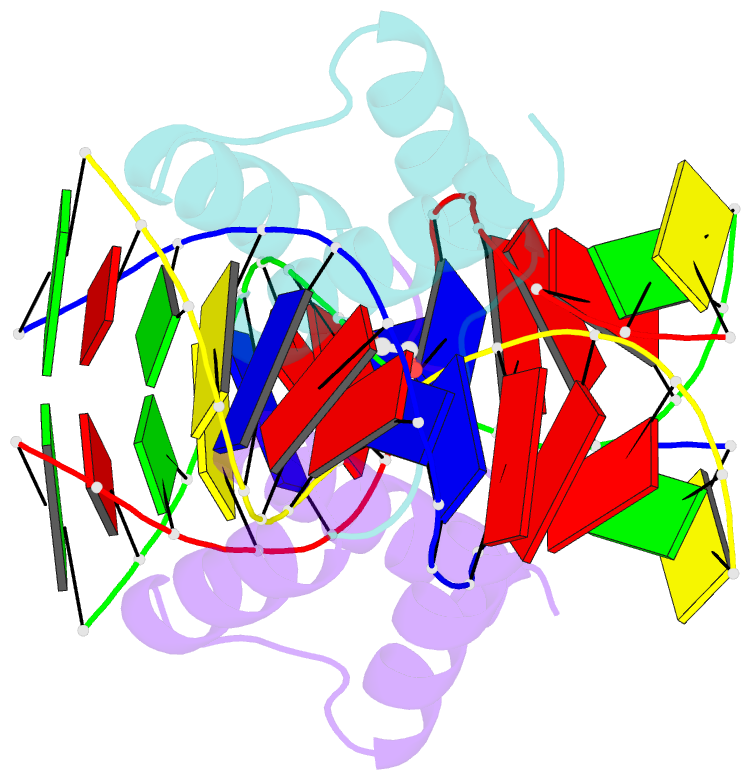
- Crystal structure of gsx2 homeodomain in complex with DNA
- Reference
- Webb JA, Farrow E, Cain B, Yuan Z, Yarawsky AE, Schoch E, Gagliani EK, Herr AB, Gebelein B, Kovall RA (2024): "Cooperative Gsx2-DNA binding requires DNA bending and a novel Gsx2 homeodomain interface." Nucleic Acids Res., 52, 7987-8002. doi: 10.1093/nar/gkae522.
- Abstract
- The conserved Gsx homeodomain (HD) transcription factors specify neural cell fates in animals from flies to mammals. Like many HD proteins, Gsx factors bind A/T-rich DNA sequences prompting the following question: How do HD factors that bind similar DNA sequences in vitro regulate specific target genes in vivo? Prior studies revealed that Gsx factors bind DNA both as a monomer on individual A/T-rich sites and as a cooperative homodimer to two sites spaced precisely 7 bp apart. However, the mechanistic basis for Gsx-DNA binding and cooperativity is poorly understood. Here, we used biochemical, biophysical, structural and modeling approaches to (i) show that Gsx factors are monomers in solution and require DNA for cooperative complex formation, (ii) define the affinity and thermodynamic binding parameters of Gsx2/DNA interactions, (iii) solve a high-resolution monomer/DNA structure that reveals that Gsx2 induces a 20° bend in DNA, (iv) identify a Gsx2 protein-protein interface required for cooperative DNA binding and (v) determine that flexible spacer DNA sequences enhance Gsx2 cooperativity on dimer sites. Altogether, our results provide a mechanistic basis for understanding the protein and DNA structural determinants that underlie cooperative DNA binding by Gsx factors.