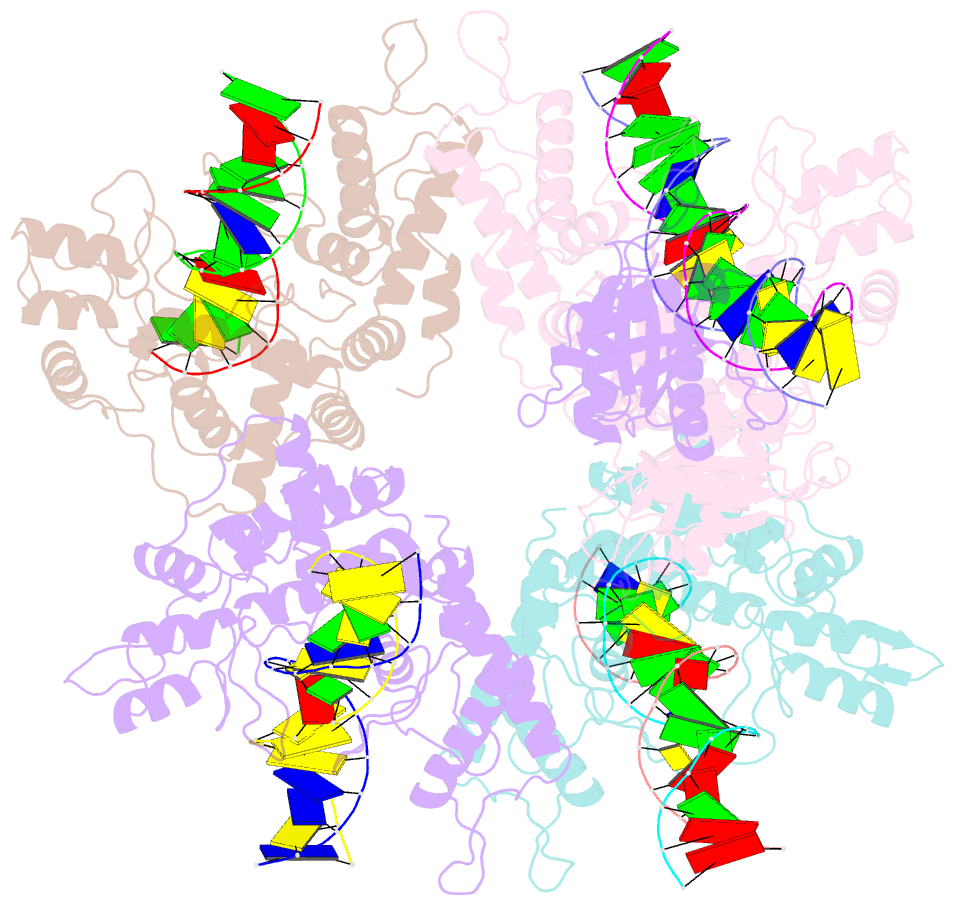
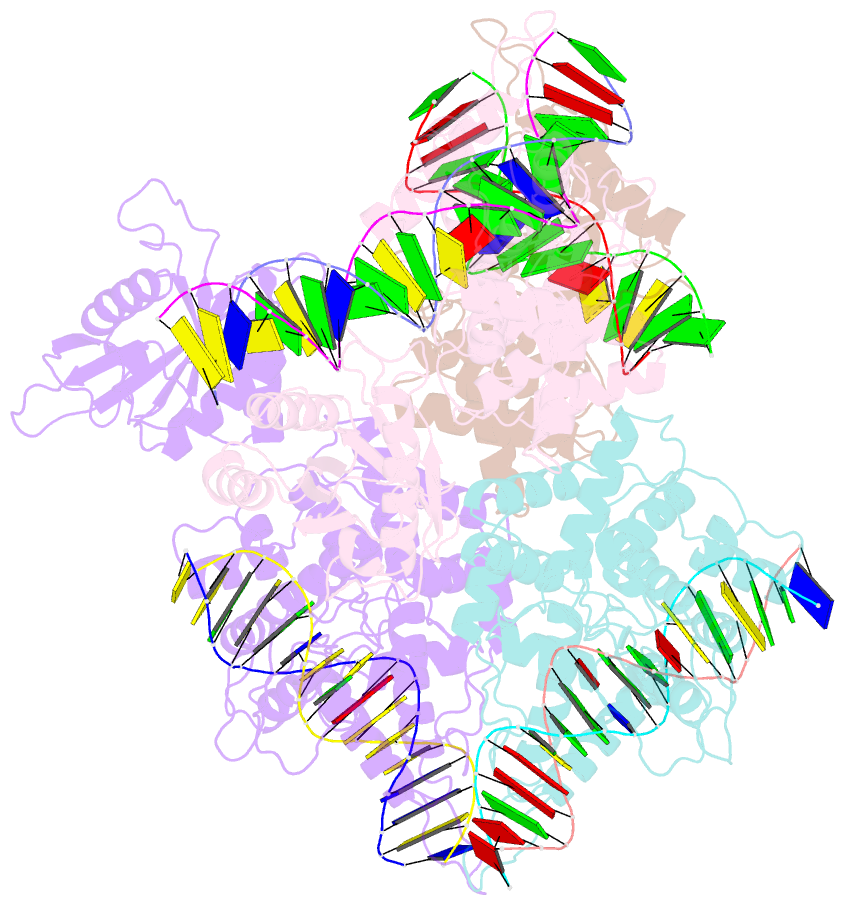
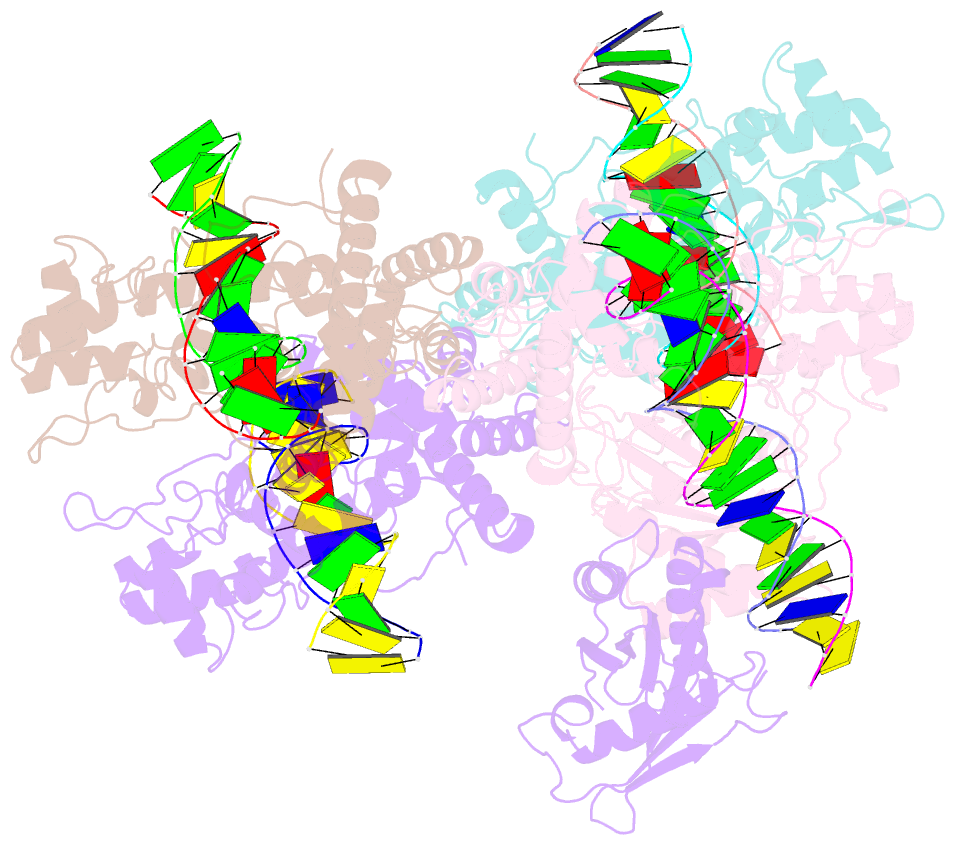
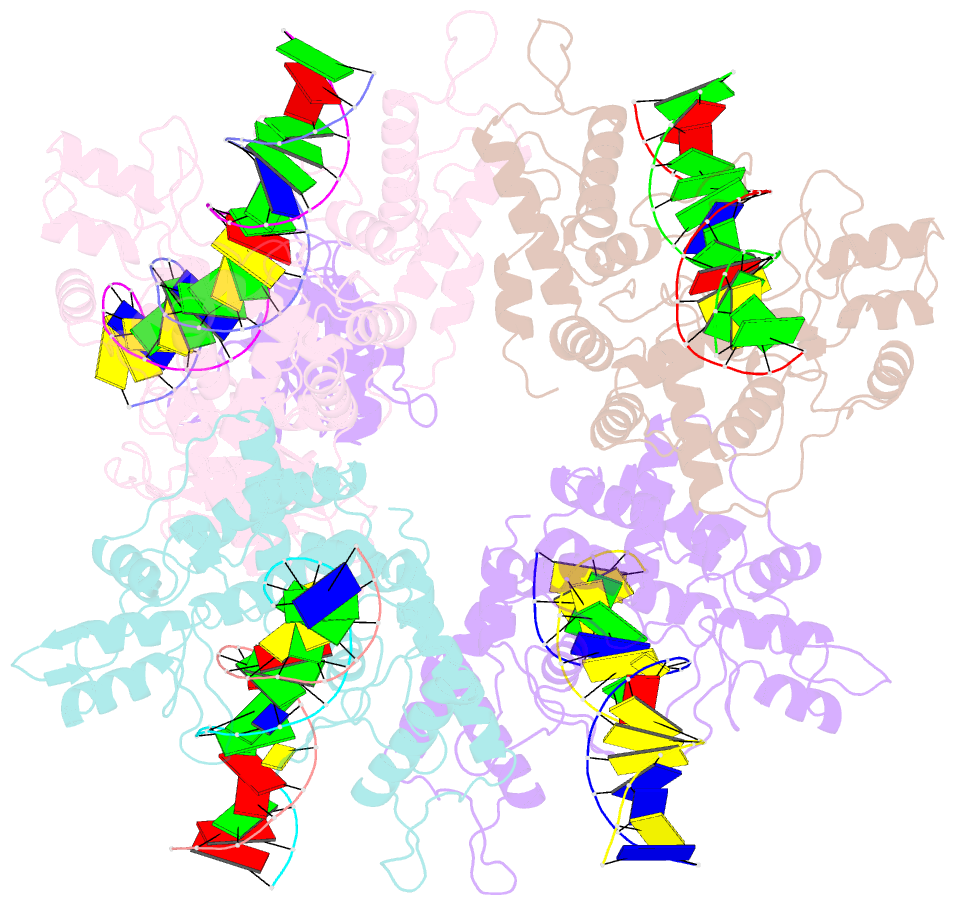
Summary information and primary citation
- PDB-id
- 8epx; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- cryo-EM (3.15 Å)
- Summary
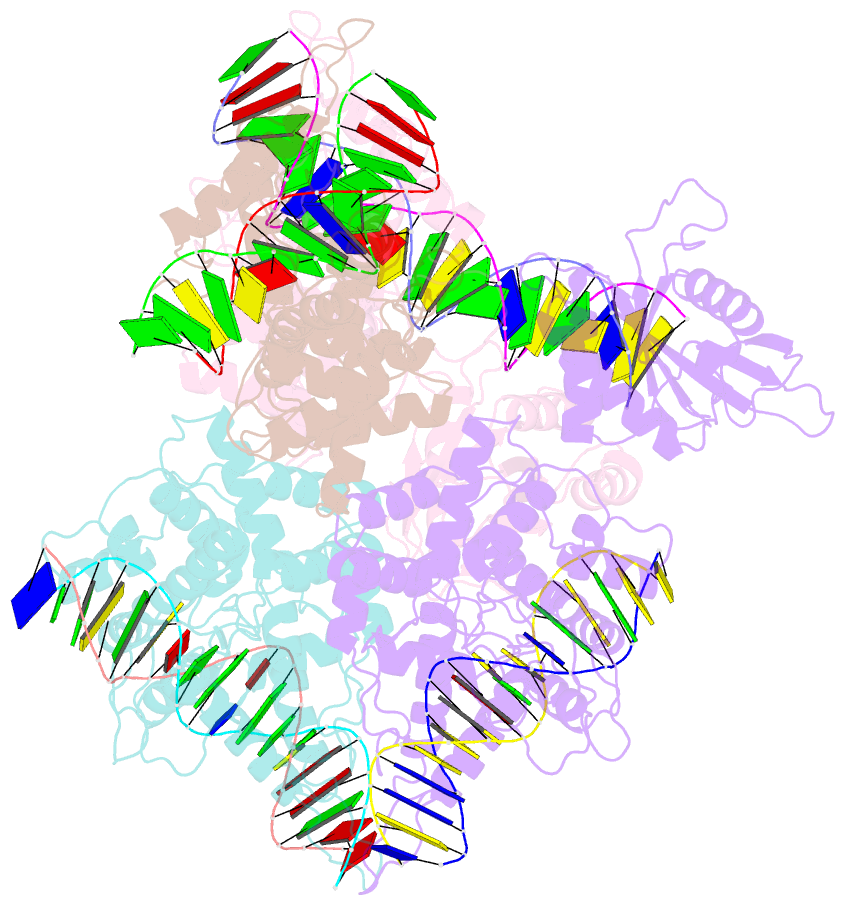
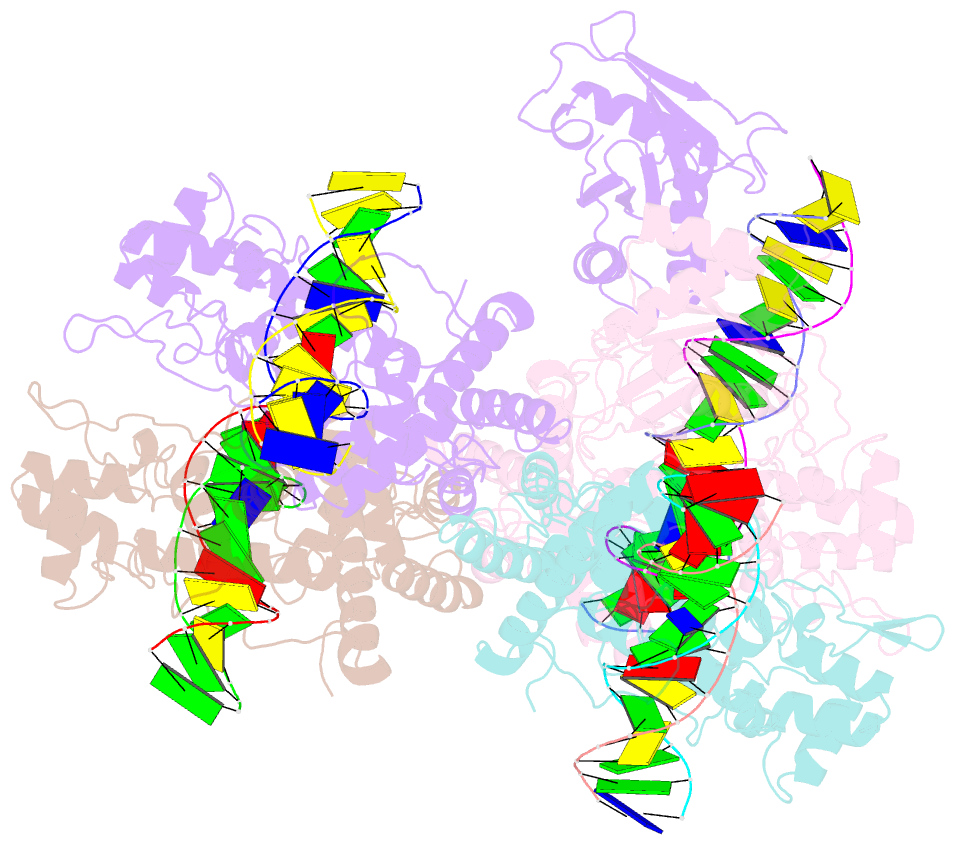
- Type iis restriction endonuclease paqci, DNA bound
- Reference
- Kennedy MA, Hosford CJ, Azumaya CM, Luyten YA, Chen M, Morgan RD, Stoddard BL (2023): "Structures, activity and mechanism of the Type IIS restriction endonuclease PaqCI." Nucleic Acids Res., 51, 4467-4487. doi: 10.1093/nar/gkad228.
- Abstract
- Type IIS restriction endonucleases contain separate DNA recognition and catalytic domains and cleave their substrates at well-defined distances outside their target sequences. They are employed in biotechnology for a variety of purposes, including the creation of gene-targeting zinc finger and TAL effector nucleases and DNA synthesis applications such as Golden Gate assembly. The most thoroughly studied Type IIS enzyme, FokI, has been shown to require multimerization and engagement with multiple DNA targets for optimal cleavage activity; however, details of how it or similar enzymes forms a DNA-bound reaction complex have not been described at atomic resolution. Here we describe biochemical analyses of DNA cleavage by the Type IIS PaqCI restriction endonuclease and a series of molecular structures in the presence and absence of multiple bound DNA targets. The enzyme displays a similar tetrameric organization of target recognition domains in the absence or presence of bound substrate, with a significant repositioning of endonuclease domains in a trapped DNA-bound complex that is poised to deliver the first of a series of double-strand breaks. PaqCI and FokI share similar structural mechanisms of DNA cleavage, but considerable differences in their domain organization and quaternary architecture, facilitating comparisons between distinct Type IIS enzymes.