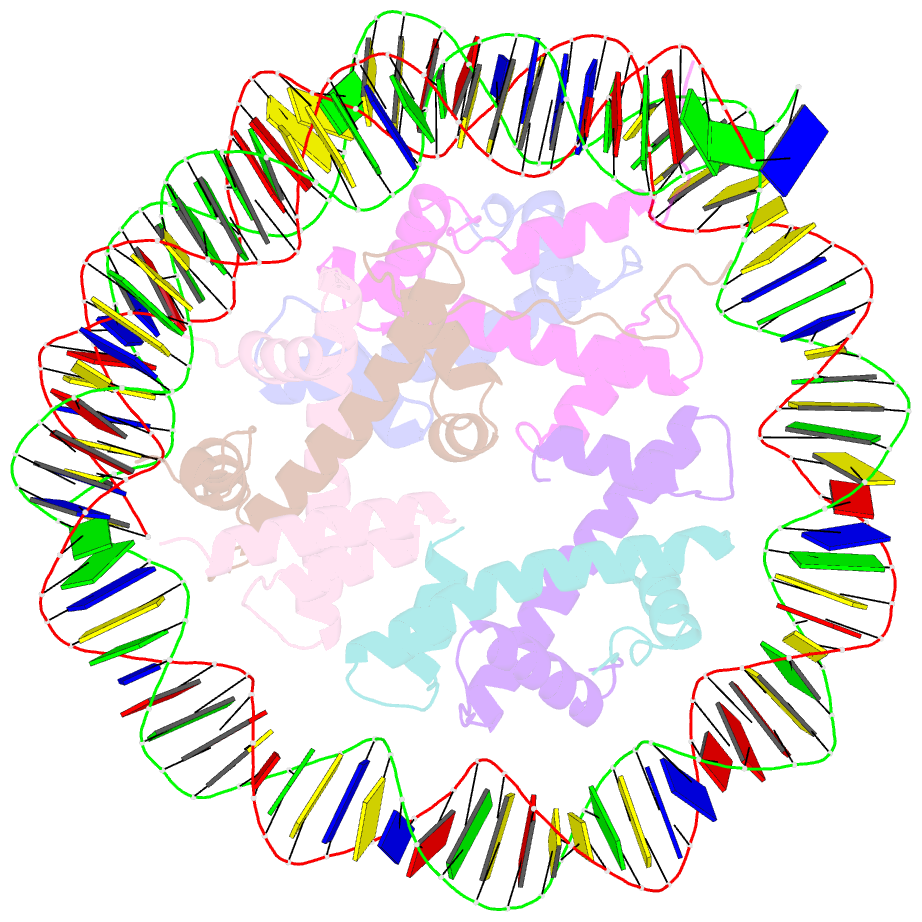
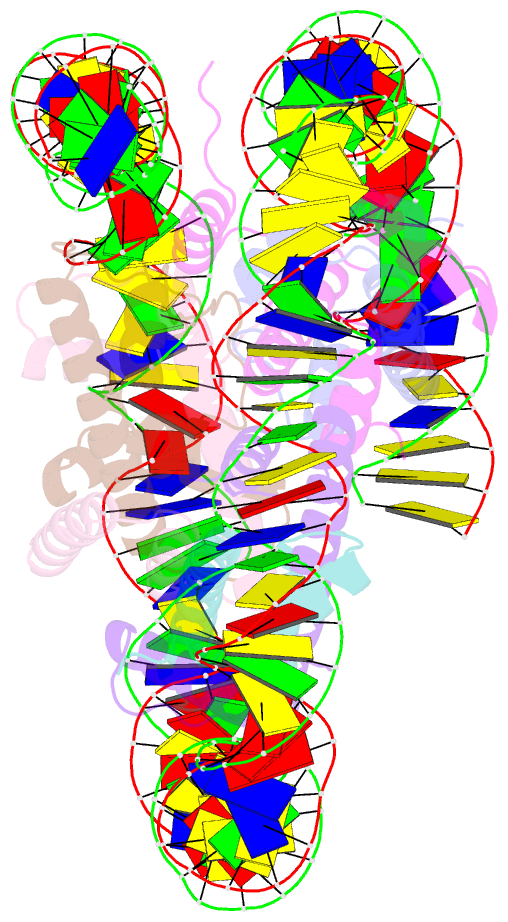
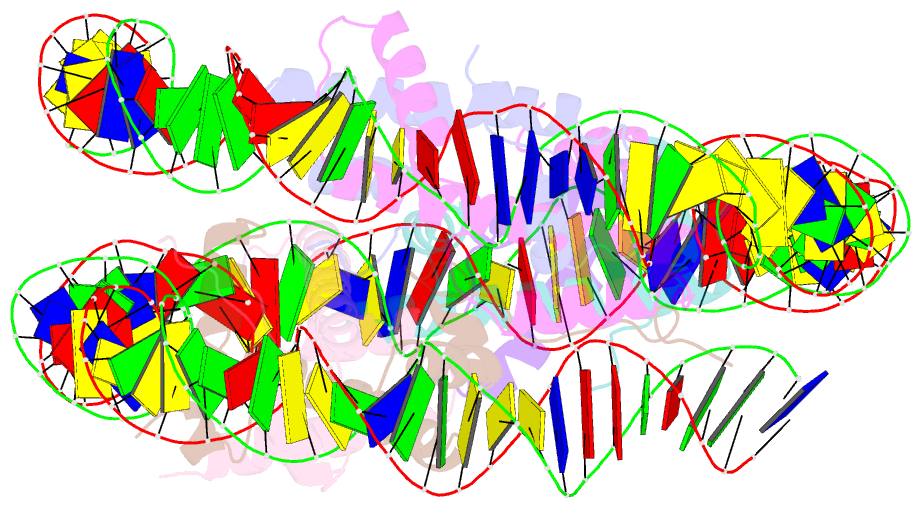
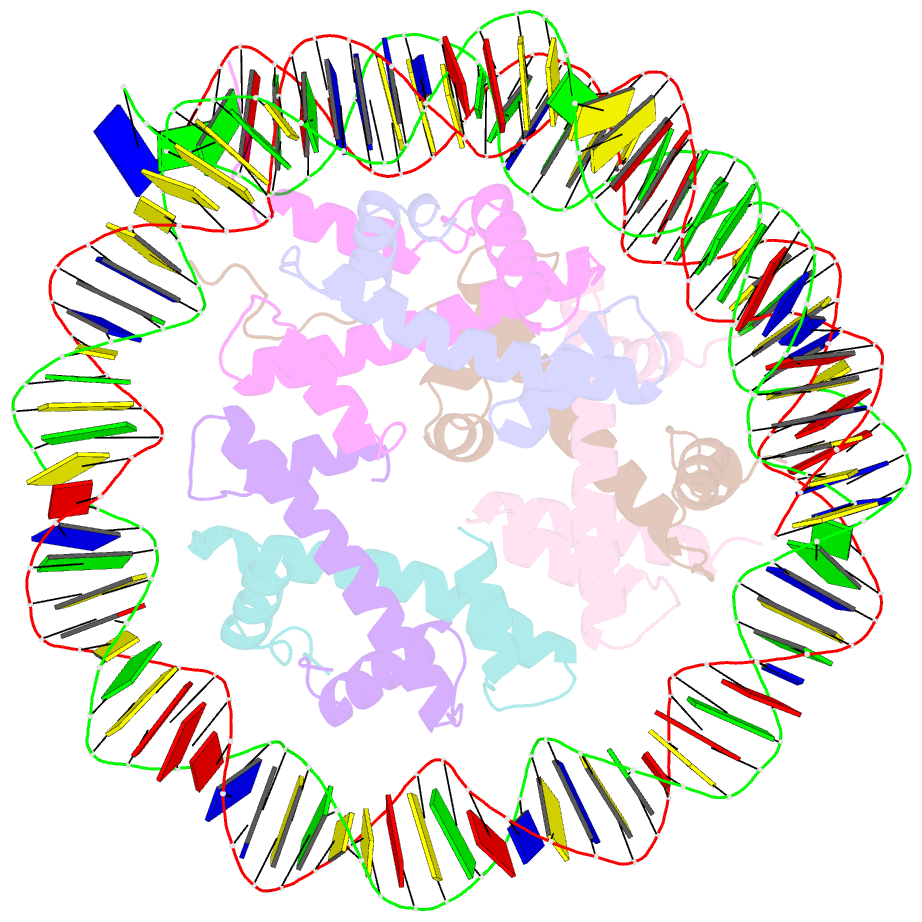
Summary information and primary citation
- PDB-id
- 8ett; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- structural protein-DNA
- Method
- cryo-EM (6.68 Å)
- Summary
- Class1 of the ino80-hexasome complex
- Reference
- Wu H, Munoz EN, Hsieh LJ, Chio US, Gourdet MA, Narlikar GJ, Cheng Y (2023): "Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility." Science, 381, 319-324. doi: 10.1126/science.adf4197.
- Abstract
- Unlike other chromatin remodelers, INO80 preferentially mobilizes hexasomes, which can form during transcription. Why INO80 prefers hexasomes over nucleosomes remains unclear. Here, we report structures of S. cerevisiae INO80 bound to a hexasome or a nucleosome. INO80 binds the two substrates in substantially different orientations. On a hexasome, INO80 places its ATPase subunit, Ino80, at superhelical location (SHL)-2, across from SHL-6/-7 as previously seen on nucleosomes. Our results suggest that INO80 action on hexasomes resembles action by other remodelers on nucleosomes, such that Ino80 is maximally active near SHL-2. The SHL-2 position also plays a critical role for nucleosome remodeling by INO80. Overall, the mechanistic adaptations used by INO80 for preferential hexasome sliding imply that sub-nucleosomal particles play considerable regulatory roles.