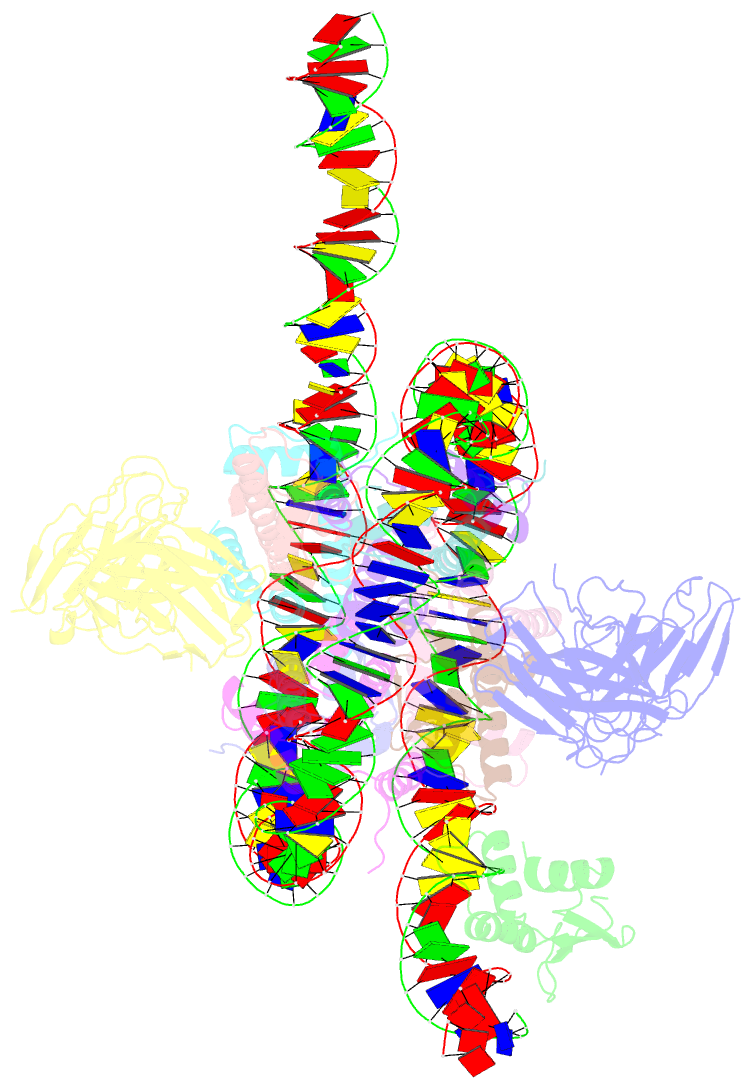
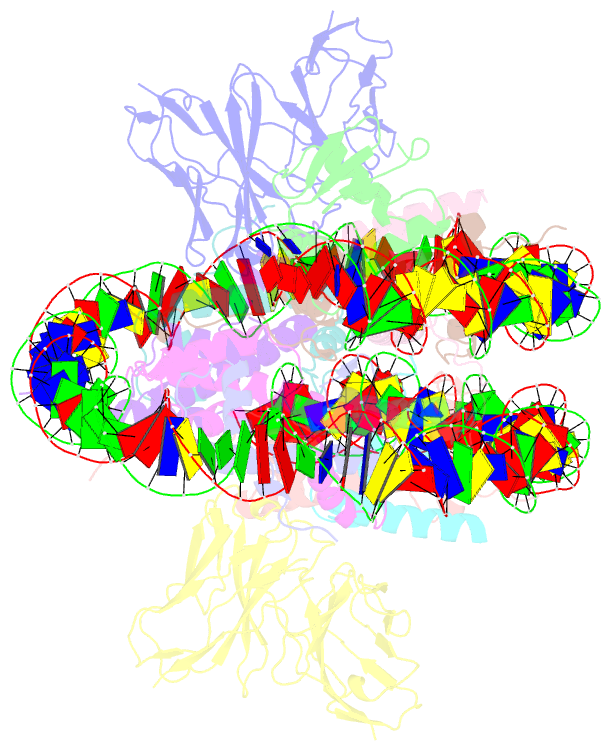
Summary information and primary citation
- PDB-id
- 8evj; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- cryo-EM (4.1 Å)
- Summary
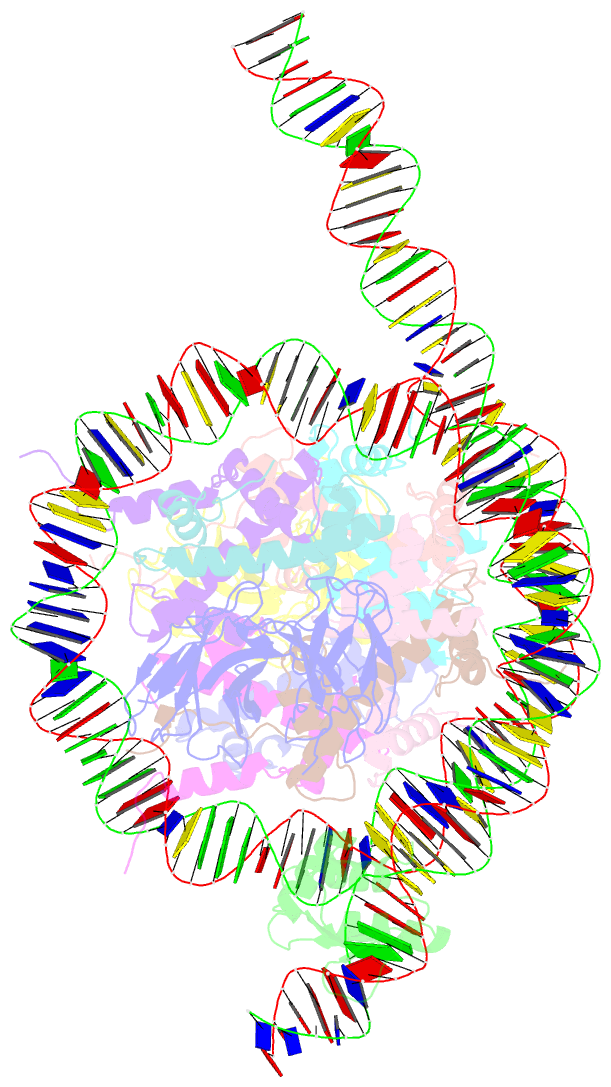
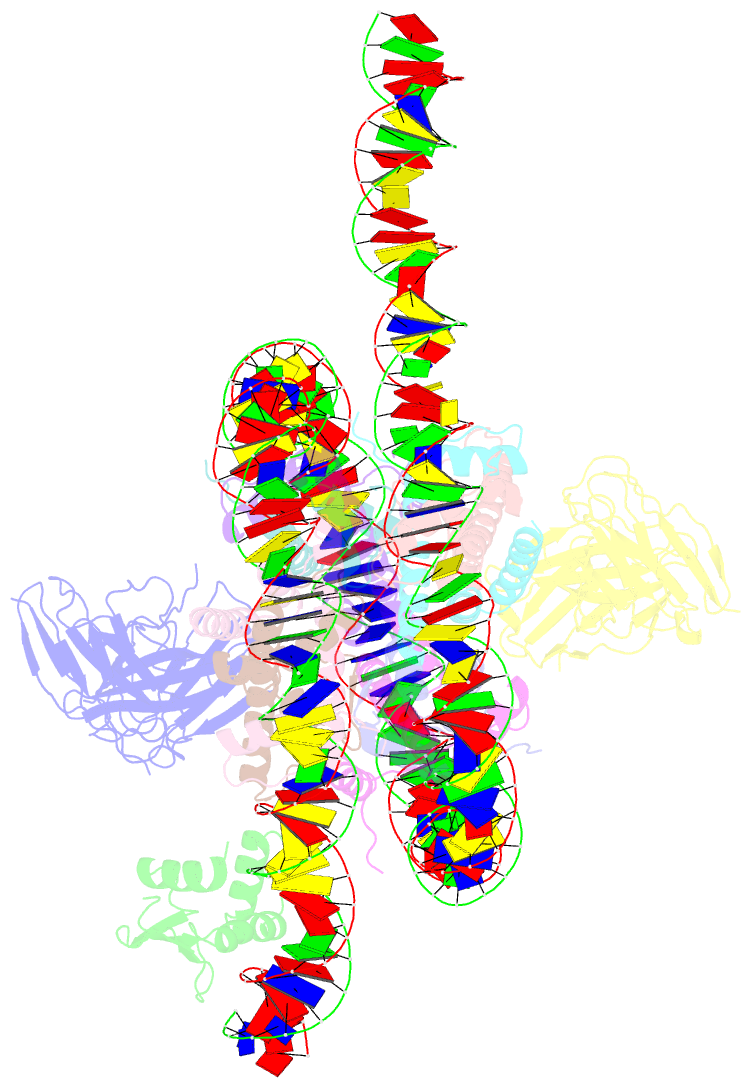
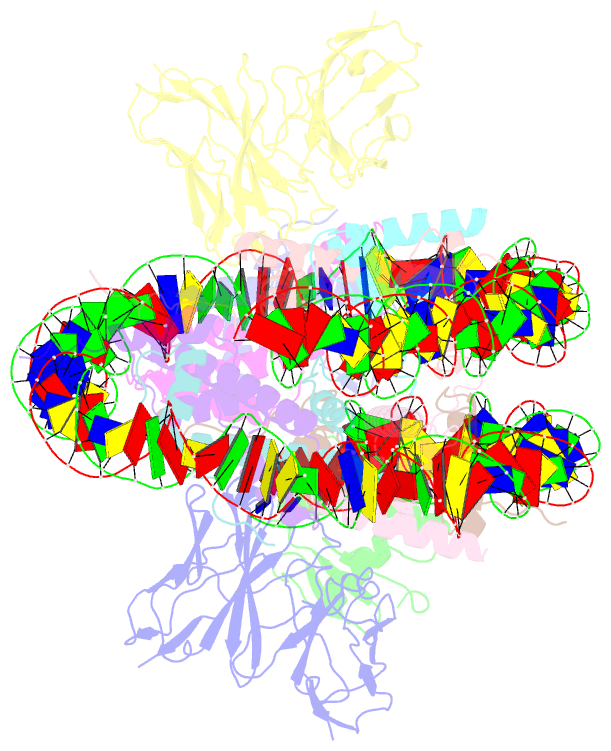
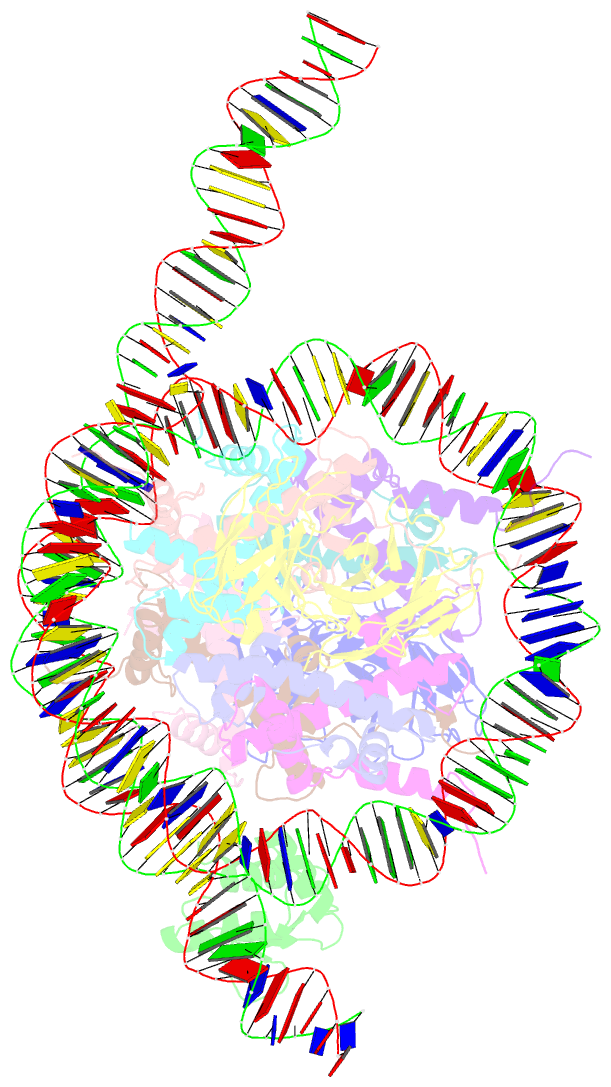
- Cx3cr1 nucleosome bound pu.1 and c-ebpa
- Reference
- Lian T, Guan R, Zhou BR, Bai Y (2024): "Structural mechanism of synergistic targeting of the CX3CR1 nucleosome by PU.1 and C/EBP alpha." Nat.Struct.Mol.Biol., 31, 633-643. doi: 10.1038/s41594-023-01189-z.
- Abstract
- Pioneer transcription factors are vital for cell fate changes. PU.1 and C/EBPα work together to regulate hematopoietic stem cell differentiation. However, how they recognize in vivo nucleosomal DNA targets remains elusive. Here we report the structures of the nucleosome containing the mouse genomic CX3CR1 enhancer DNA and its complexes with PU.1 alone and with both PU.1 and the C/EBPα DNA binding domain. Our structures reveal that PU.1 binds the DNA motif at the exit linker, shifting 17 bp of DNA into the core region through interactions with H2A, unwrapping ~20 bp of nucleosomal DNA. C/EBPα binding, aided by PU.1's repositioning, unwraps ~25 bp of entry DNA. The PU.1 Q218H mutation, linked to acute myeloid leukemia, disrupts PU.1-H2A interactions. PU.1 and C/EBPα jointly displace linker histone H1 and open the H1-condensed nucleosome array. Our study unveils how two pioneer factors can work cooperatively to open closed chromatin by altering DNA positioning in the nucleosome.