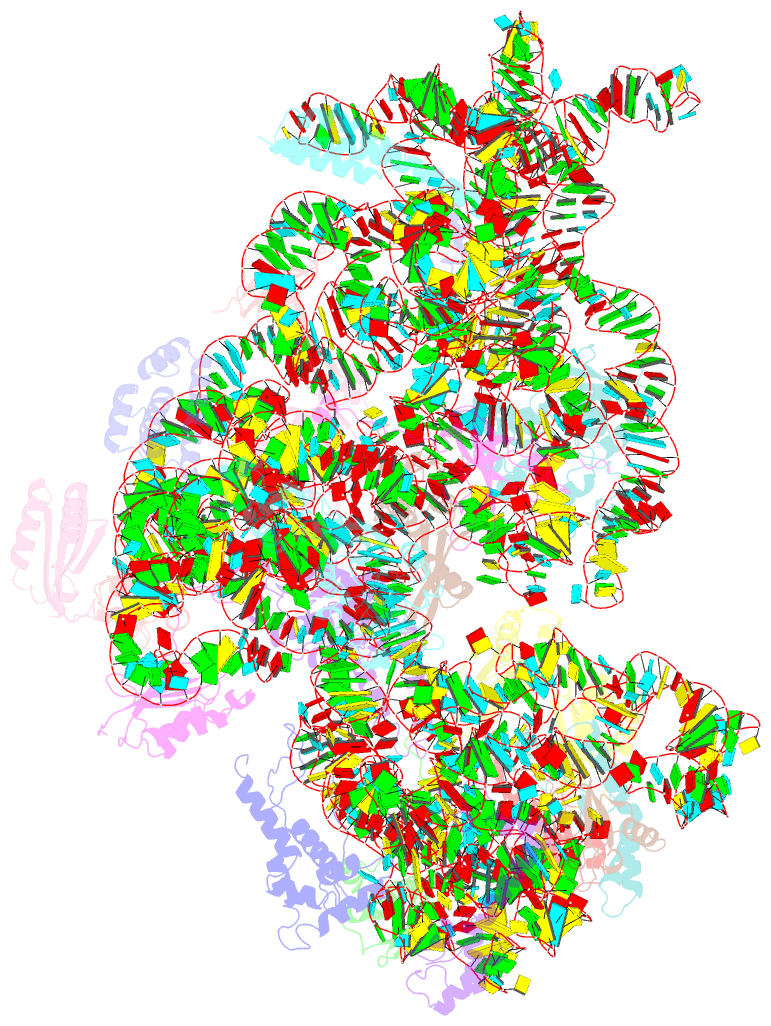
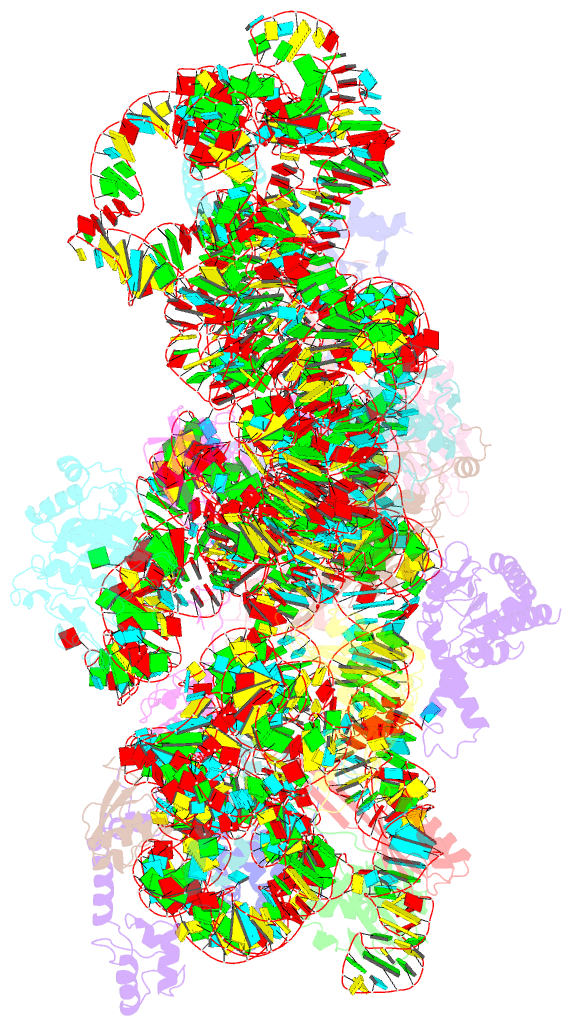
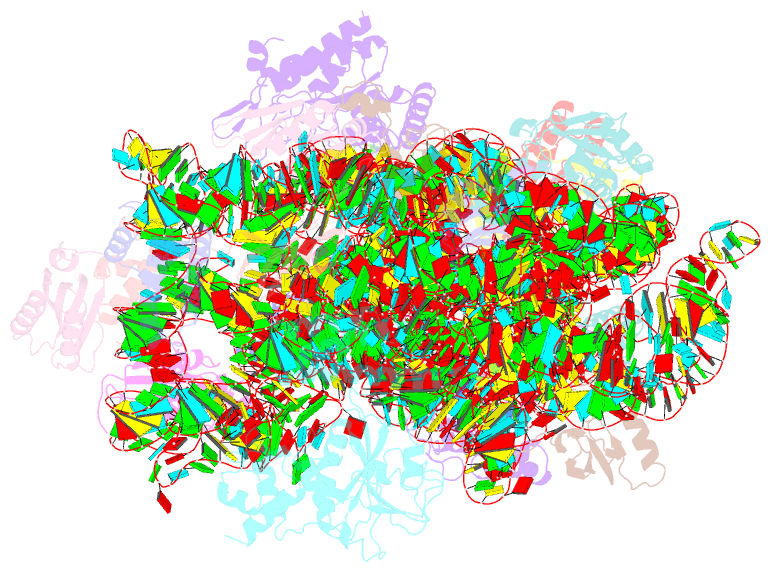
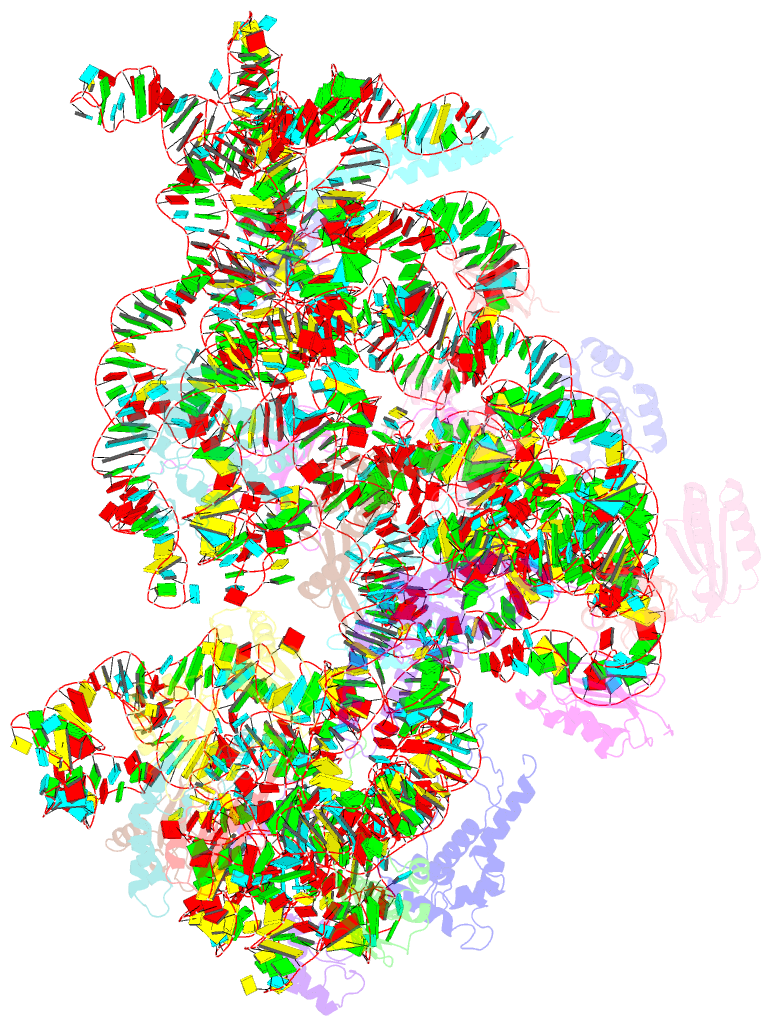
Summary information and primary citation
- PDB-id
- 8eyt; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ribosome
- Method
- cryo-EM (2.8 Å)
- Summary
- 30s_delta_ksga+ksga complex
- Reference
- Sun J, Kinman LF, Jahagirdar D, Ortega J, Davis JH (2023): "KsgA facilitates ribosomal small subunit maturation by proofreading a key structural lesion." Nat.Struct.Mol.Biol., 30, 1468-1480. doi: 10.1038/s41594-023-01078-5.
- Abstract
- Ribosome assembly is orchestrated by many assembly factors, including ribosomal RNA methyltransferases, whose precise role is poorly understood. Here, we leverage the power of cryo-EM and machine learning to discover that the E. coli methyltransferase KsgA performs a 'proofreading' function in the assembly of the small ribosomal subunit by recognizing and partially disassembling particles that have matured but are not competent for translation. We propose that this activity allows inactive particles an opportunity to reassemble into an active state, thereby increasing overall assembly fidelity. Detailed structural quantifications in our datasets additionally enabled the expansion of the Nomura assembly map to highlight rRNA helix and r-protein interdependencies, detailing how the binding and docking of these elements are tightly coupled. These results have wide-ranging implications for our understanding of the quality-control mechanisms governing ribosome biogenesis and showcase the power of heterogeneity analysis in cryo-EM to unveil functionally relevant information in biological systems.