Summary information and primary citation
- PDB-id
- 8f86; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- gene regulation
- Method
- cryo-EM (3.1 Å)
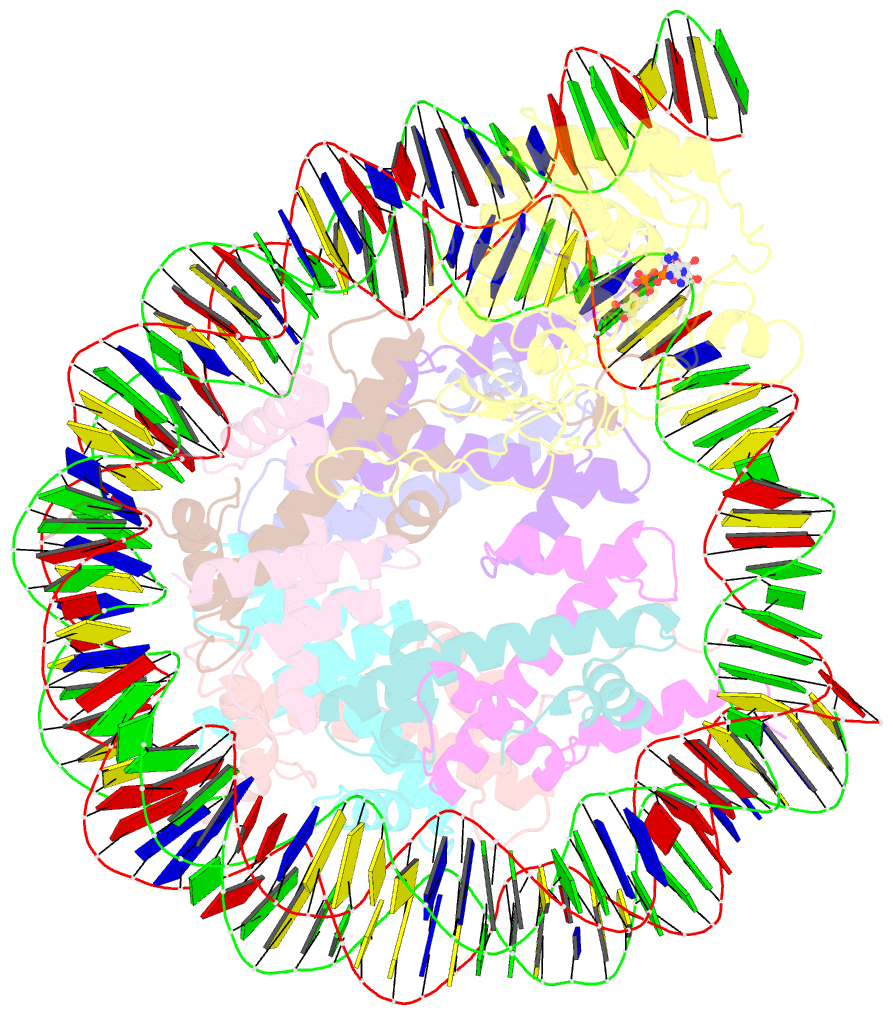
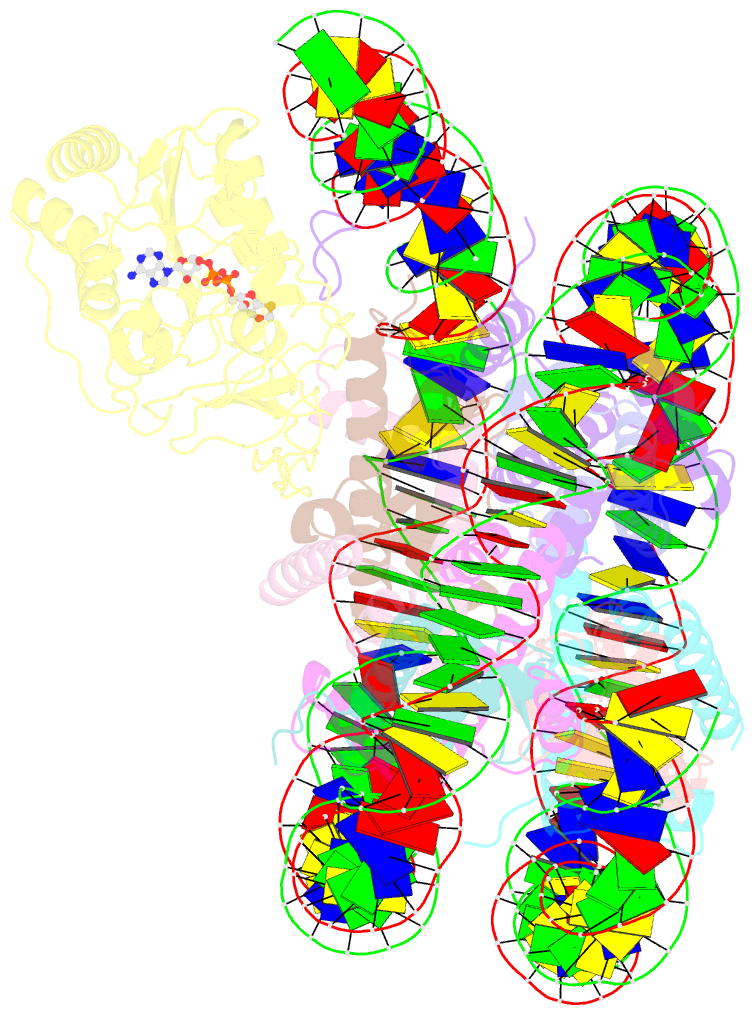
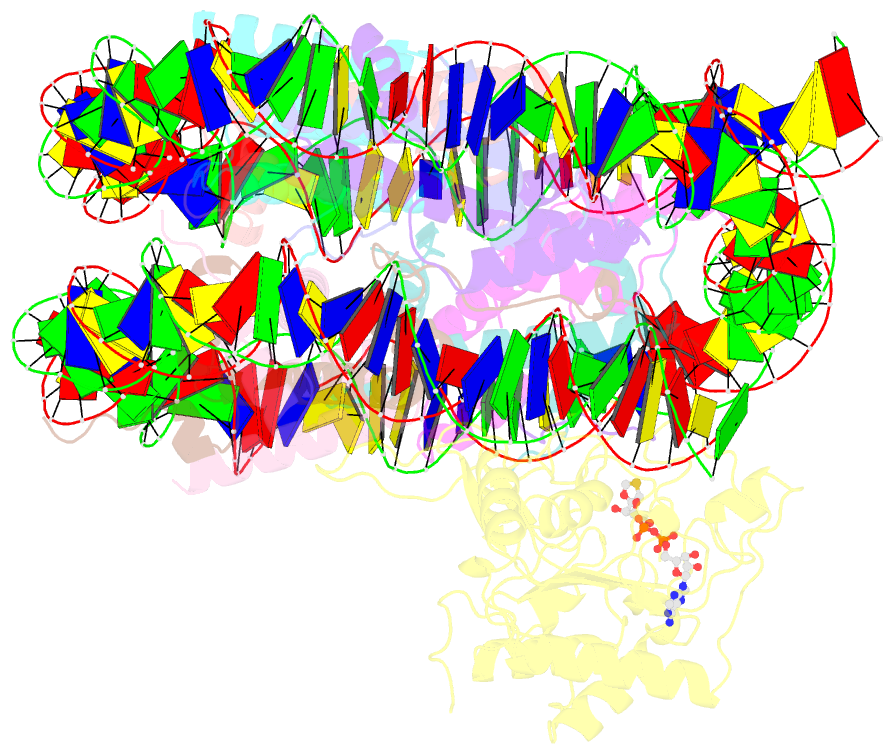
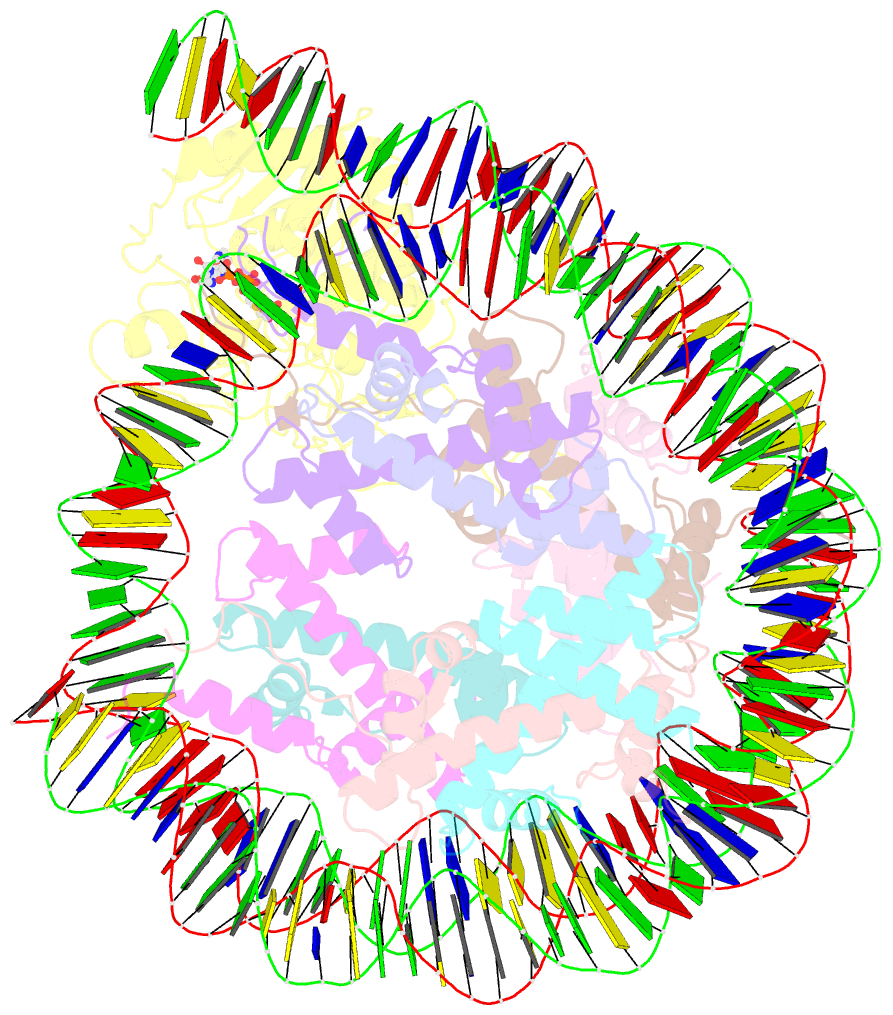
- Summary
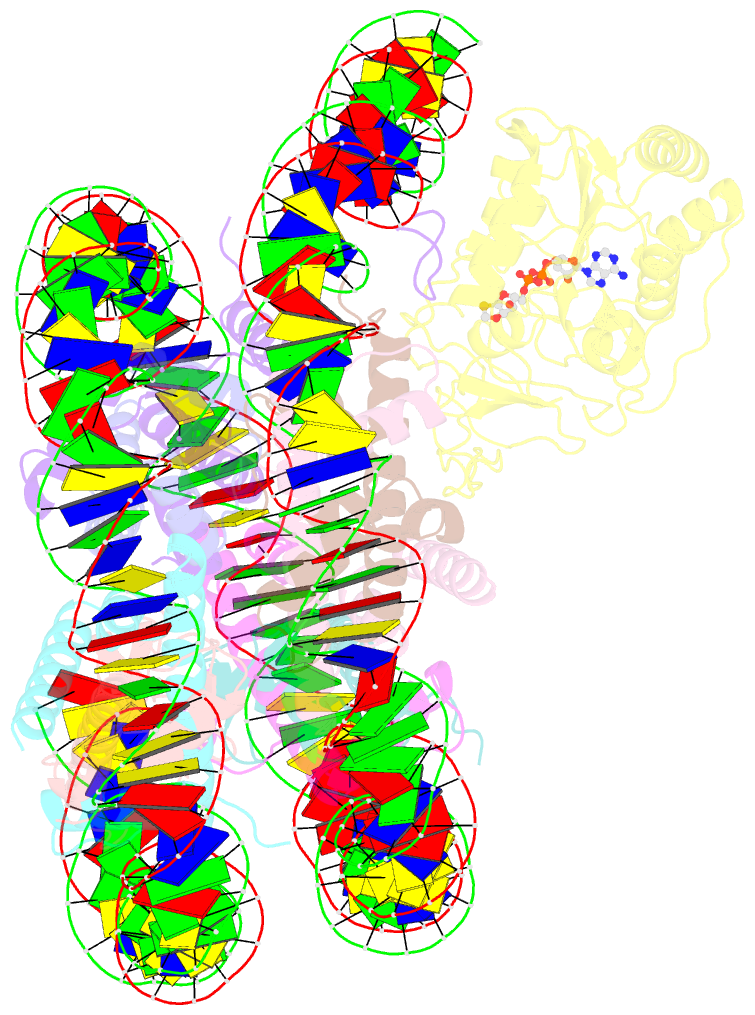
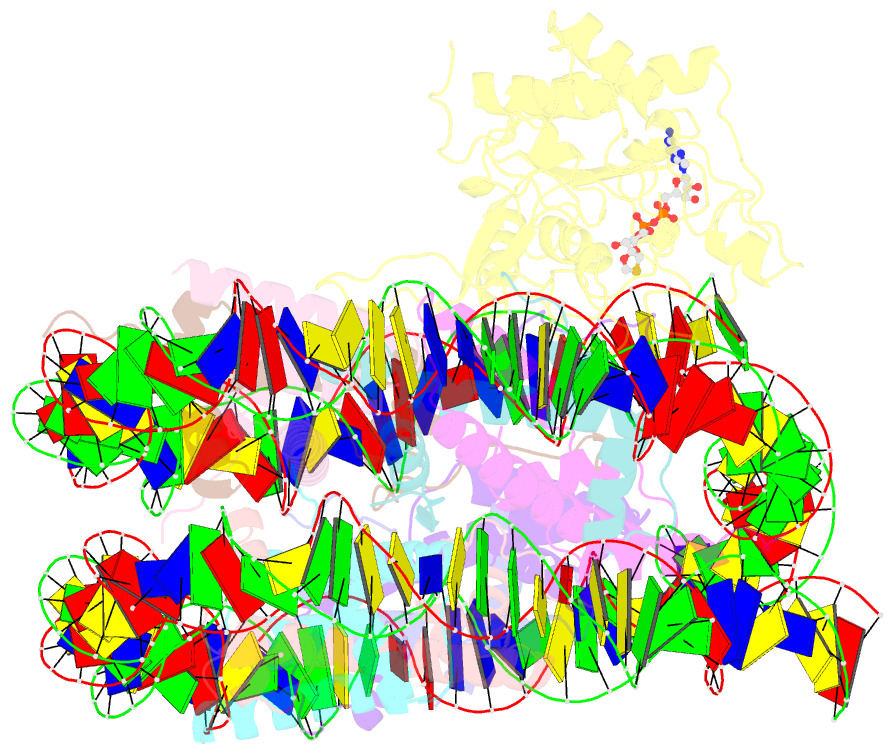
- Sirt6 bound to an h3k9ac nucleosome
- Reference
- Wang ZA, Markert JW, Whedon SD, Yapa Abeywardana M, Lee K, Jiang H, Suarez C, Lin H, Farnung L, Cole PA (2023): "Structural Basis of Sirtuin 6-Catalyzed Nucleosome Deacetylation." J.Am.Chem.Soc., 145, 6811-6822. doi: 10.1021/jacs.2c13512.
- Abstract
- The reversible acetylation of histone lysine residues is controlled by the action of acetyltransferases and deacetylases (HDACs), which regulate chromatin structure and gene expression. The sirtuins are a family of NAD-dependent HDAC enzymes, and one member, sirtuin 6 (Sirt6), influences DNA repair, transcription, and aging. Here, we demonstrate that Sirt6 is efficient at deacetylating several histone H3 acetylation sites, including its canonical site Lys9, in the context of nucleosomes but not free acetylated histone H3 protein substrates. By installing a chemical warhead at the Lys9 position of histone H3, we trap a catalytically poised Sirt6 in complex with a nucleosome and employ this in cryo-EM structural analysis. The structure of Sirt6 bound to a nucleosome reveals extensive interactions between distinct segments of Sirt6 and the H2A/H2B acidic patch and nucleosomal DNA, which accounts for the rapid deacetylation of nucleosomal H3 sites and the disfavoring of histone H2B acetylation sites. These findings provide a new framework for understanding how HDACs target and regulate chromatin.