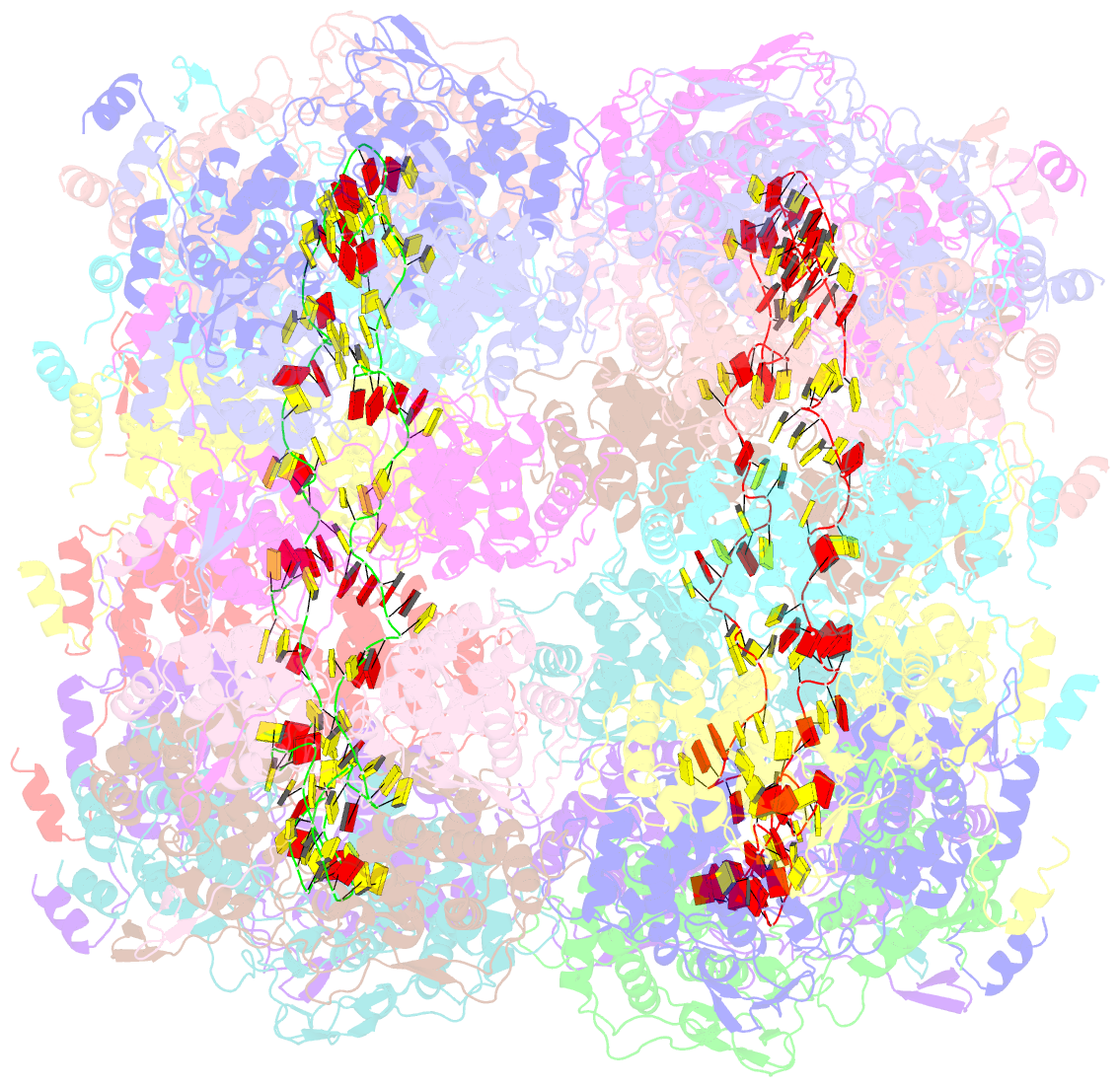
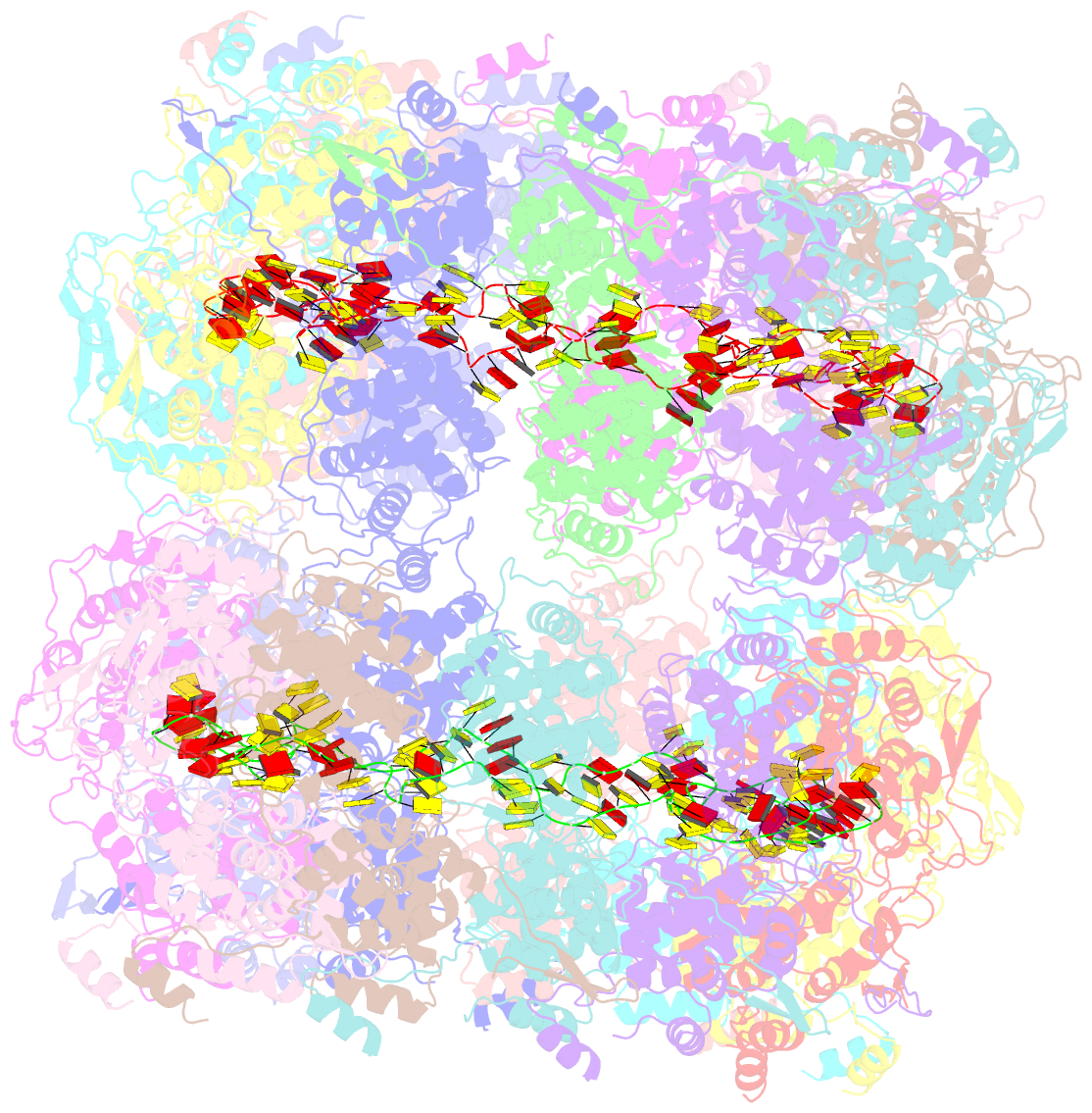
Summary information and primary citation
- PDB-id
- 8ffr; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein
- Method
- X-ray (3.49 Å)
- Summary
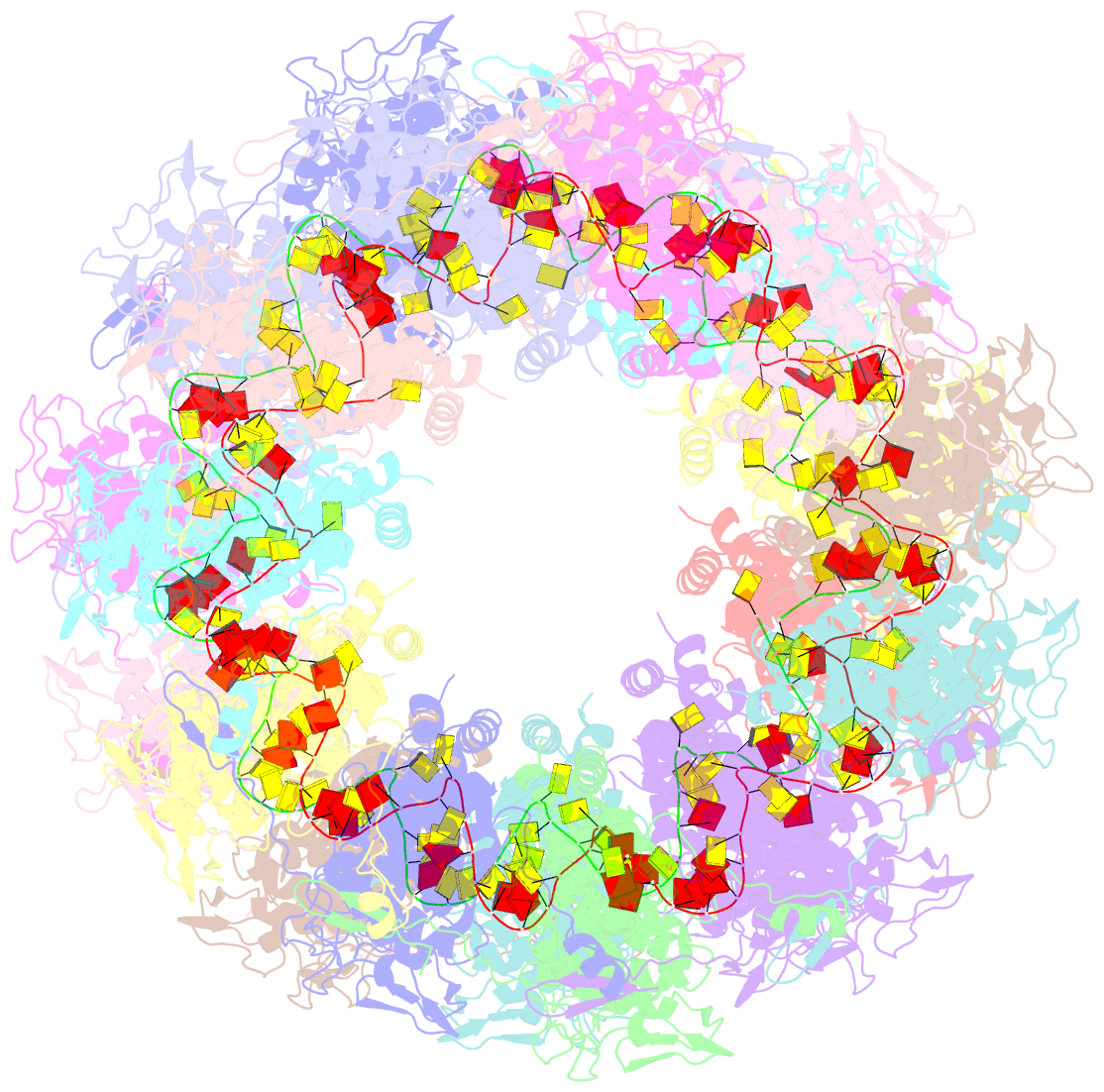
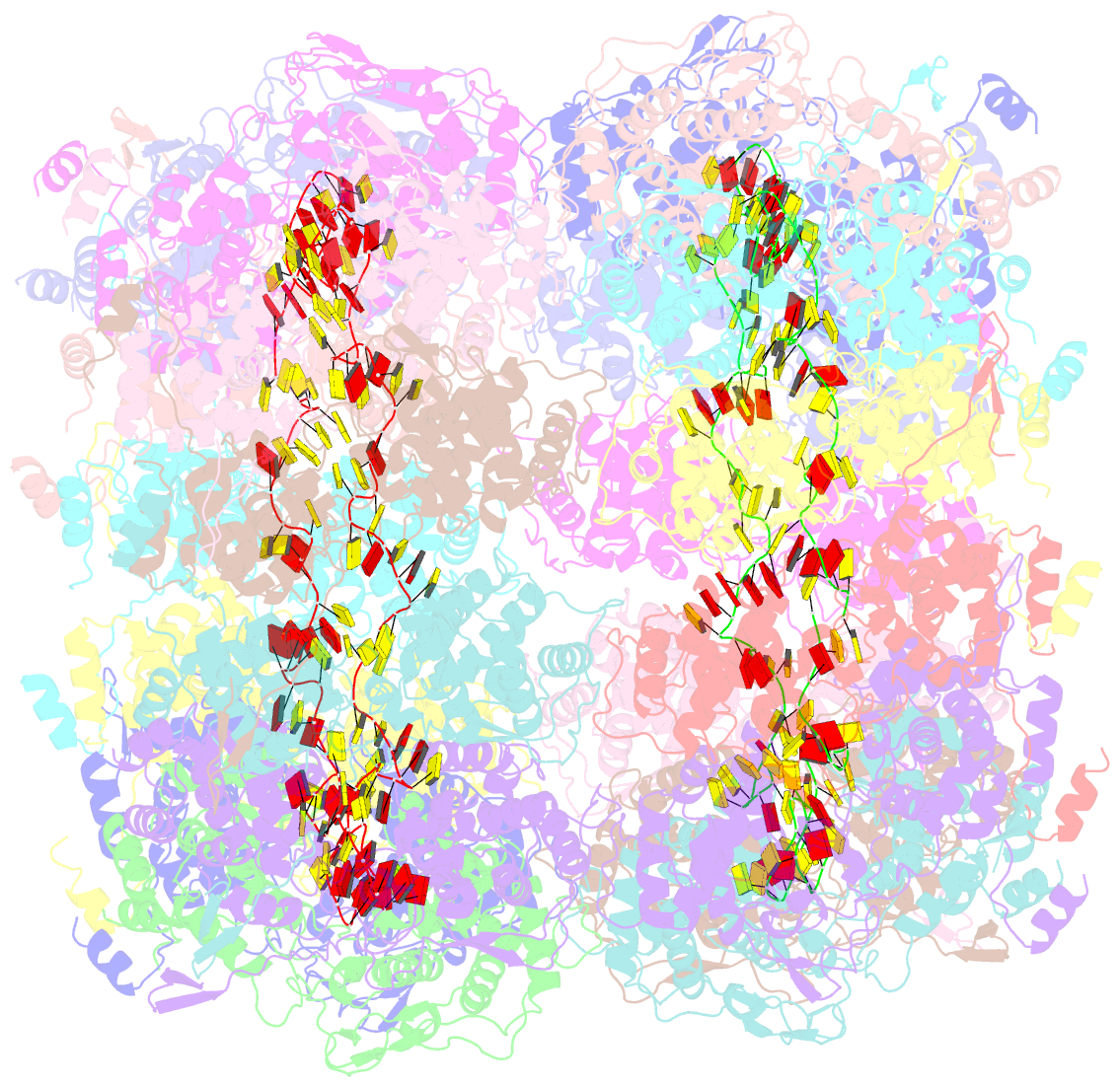
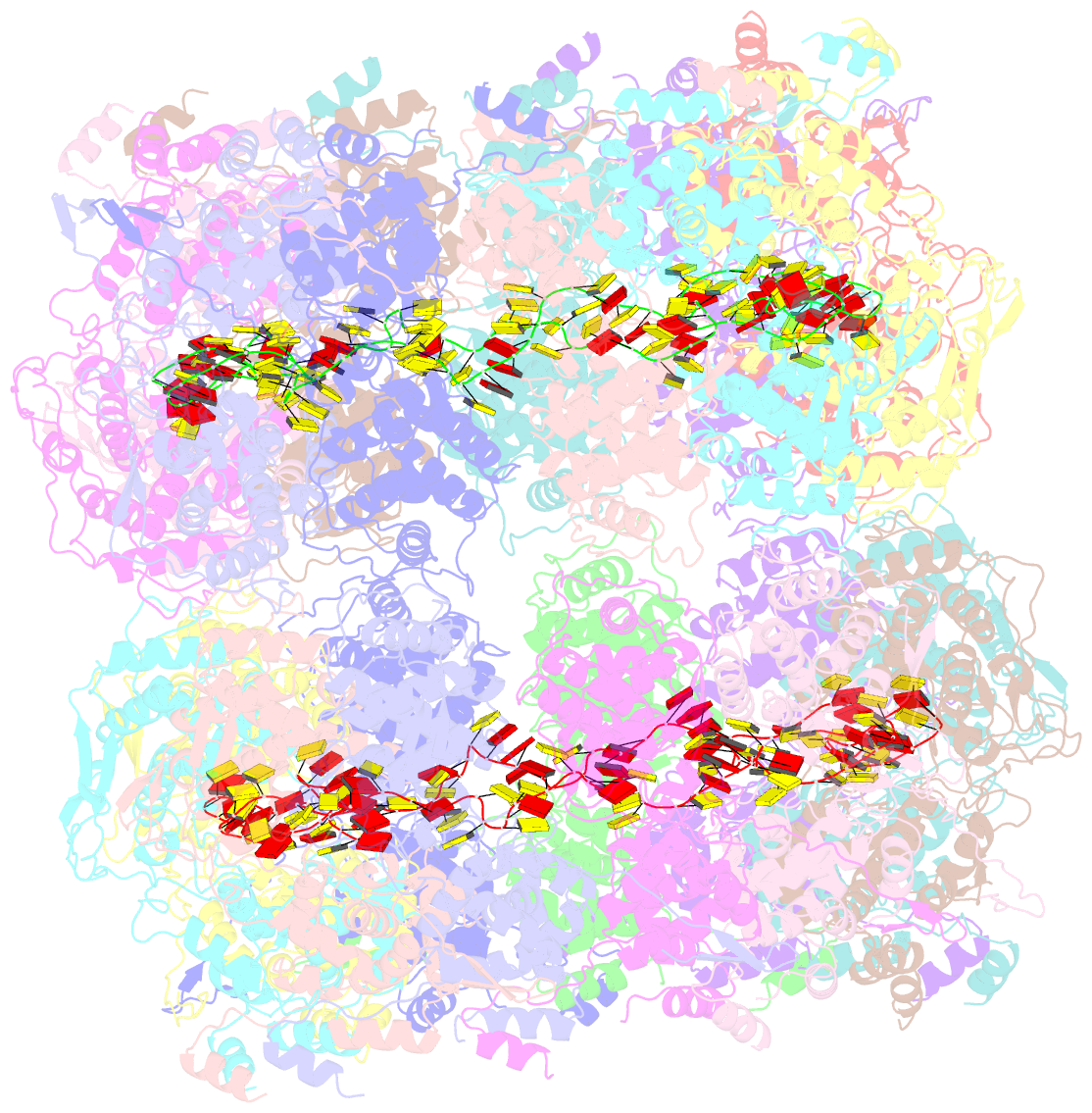
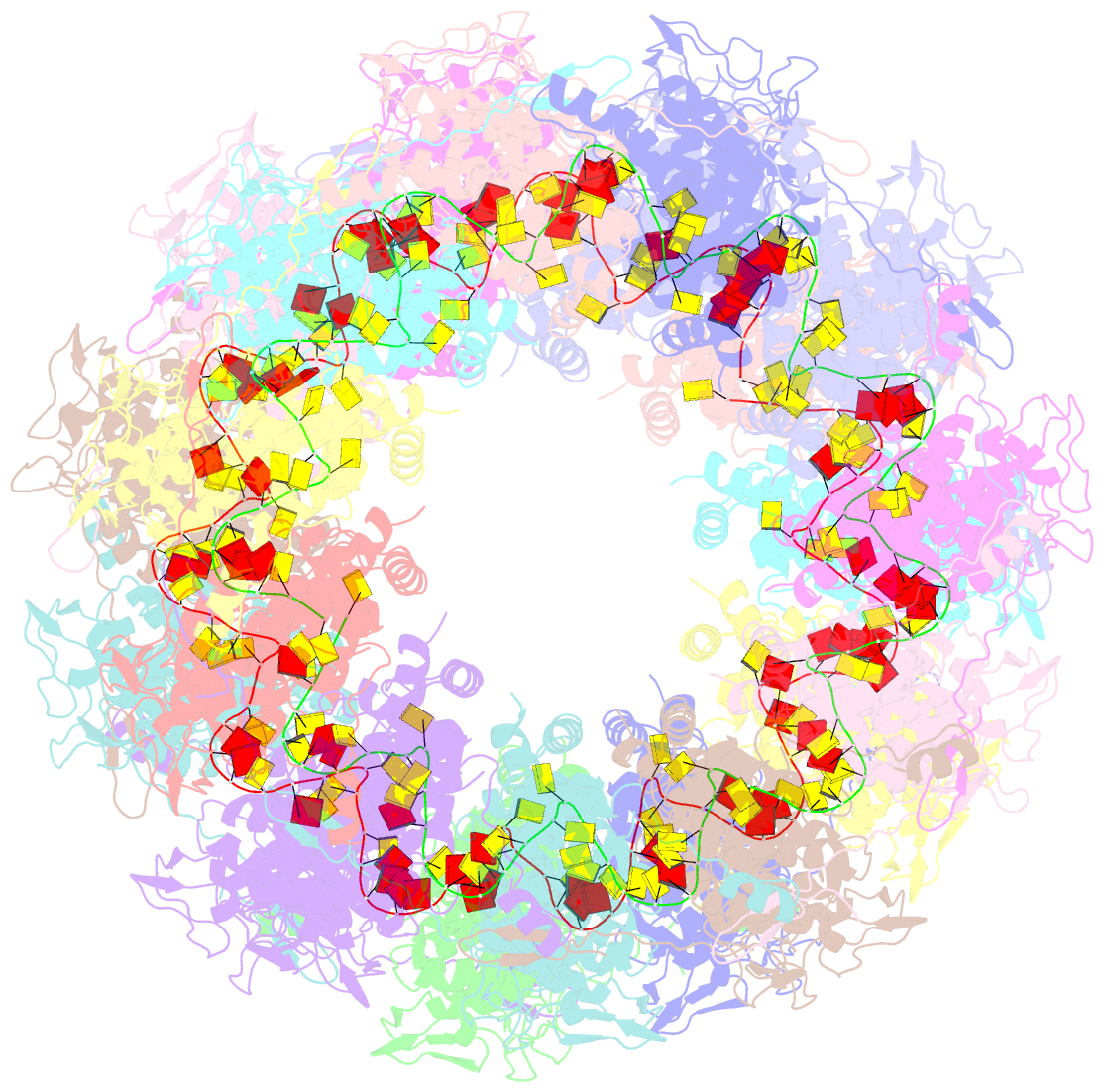
- Revised structure of the rabies virus nucleoprotein-RNA complex
- Reference
- Gerard FCA, Bourhis JM, Mas C, Branchard A, Vu DD, Varhoshkova S, Leyrat C, Jamin M (2022): "Structure and Dynamics of the Unassembled Nucleoprotein of Rabies Virus in Complex with Its Phosphoprotein Chaperone Module." Viruses, 14. doi: 10.3390/v14122813.
- Abstract
- As for all non-segmented negative RNA viruses, rabies virus has its genome packaged in a linear assembly of nucleoprotein (N), named nucleocapsid. The formation of new nucleocapsids during virus replication in cells requires the production of soluble N protein in complex with its phosphoprotein (P) chaperone. In this study, we reconstituted a soluble heterodimeric complex between an armless N protein of rabies virus (RABV), lacking its N-terminal subdomain (NNT-ARM), and a peptide encompassing the N0 chaperon module of the P protein. We showed that the chaperone module undergoes a disordered-order transition when it assembles with N0 and measured an affinity in the low nanomolar range using a competition assay. We solved the crystal structure of the complex at a resolution of 2.3 Å, unveiling the details of the conserved interfaces. MD simulations showed that both the chaperon module of P and RNA-mediated polymerization reduced the ability of the RNA binding cavity to open and close. Finally, by reconstituting a complex with full-length P protein, we demonstrated that each P dimer could independently chaperon two N0 molecules.