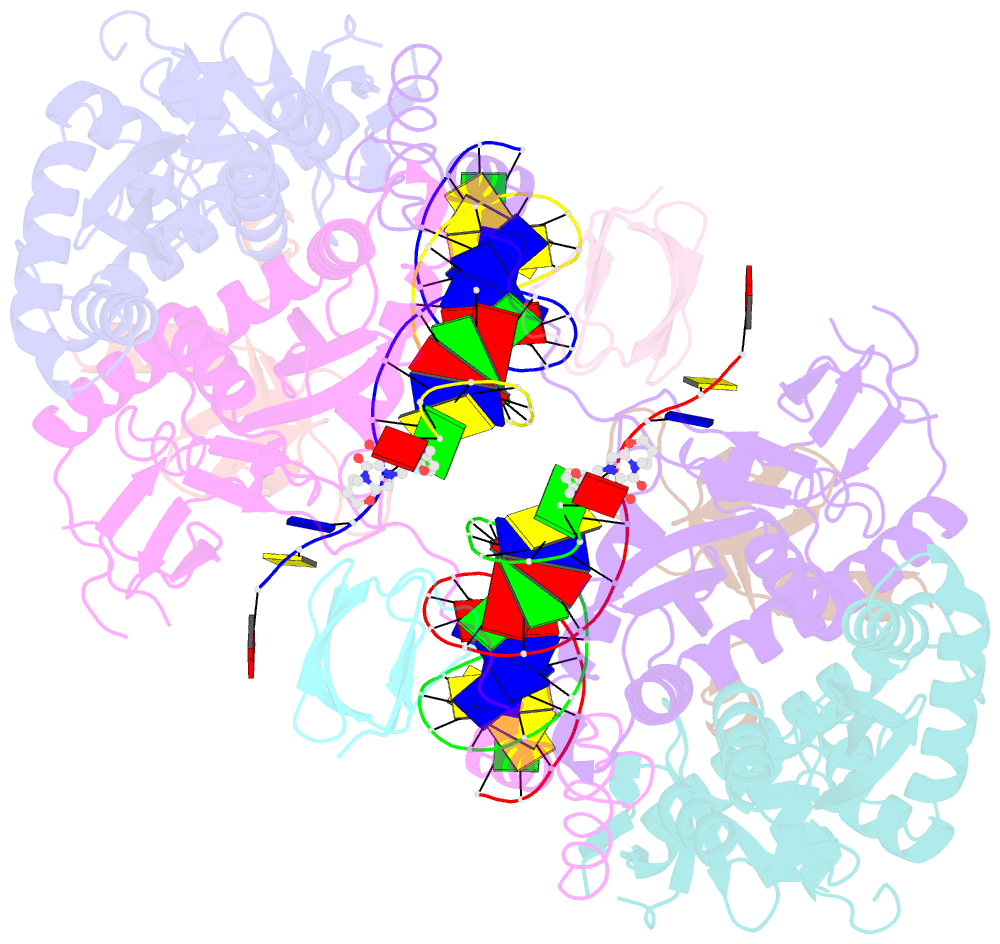
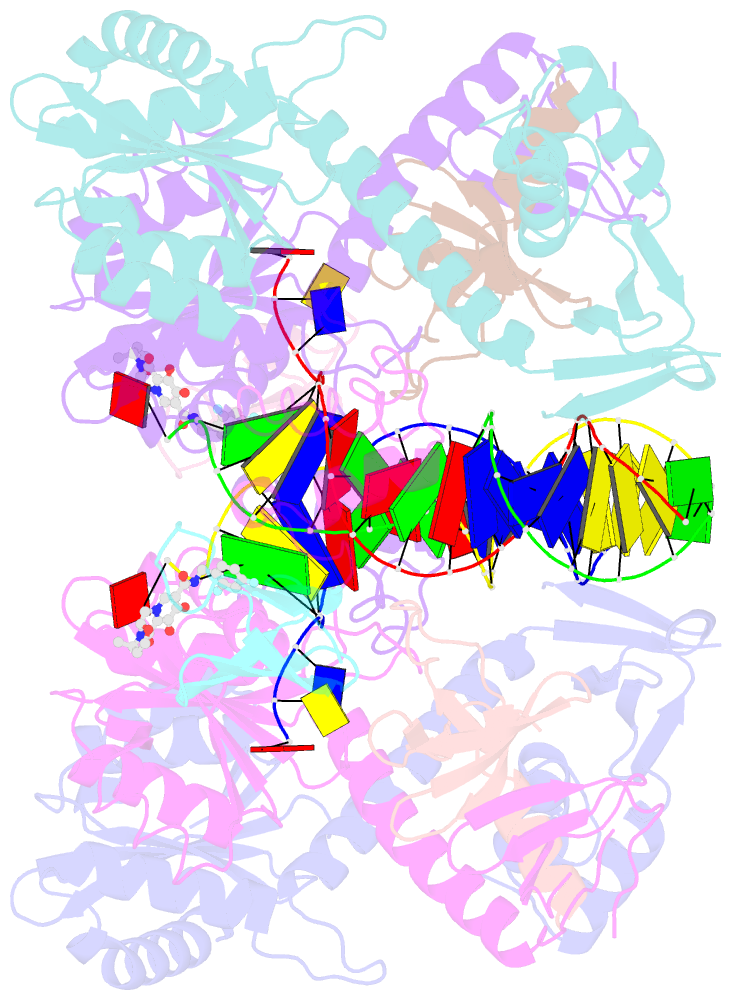
Summary information and primary citation
- PDB-id
- 8fng; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein-DNA-inhibitor
- Method
- cryo-EM (2.2 Å)
- Summary
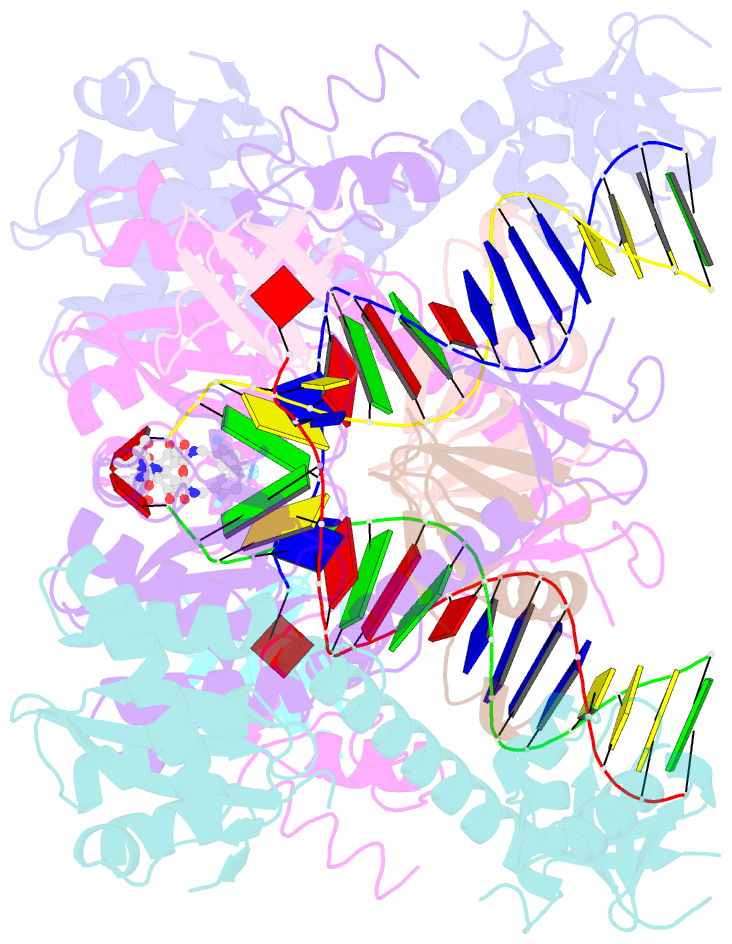
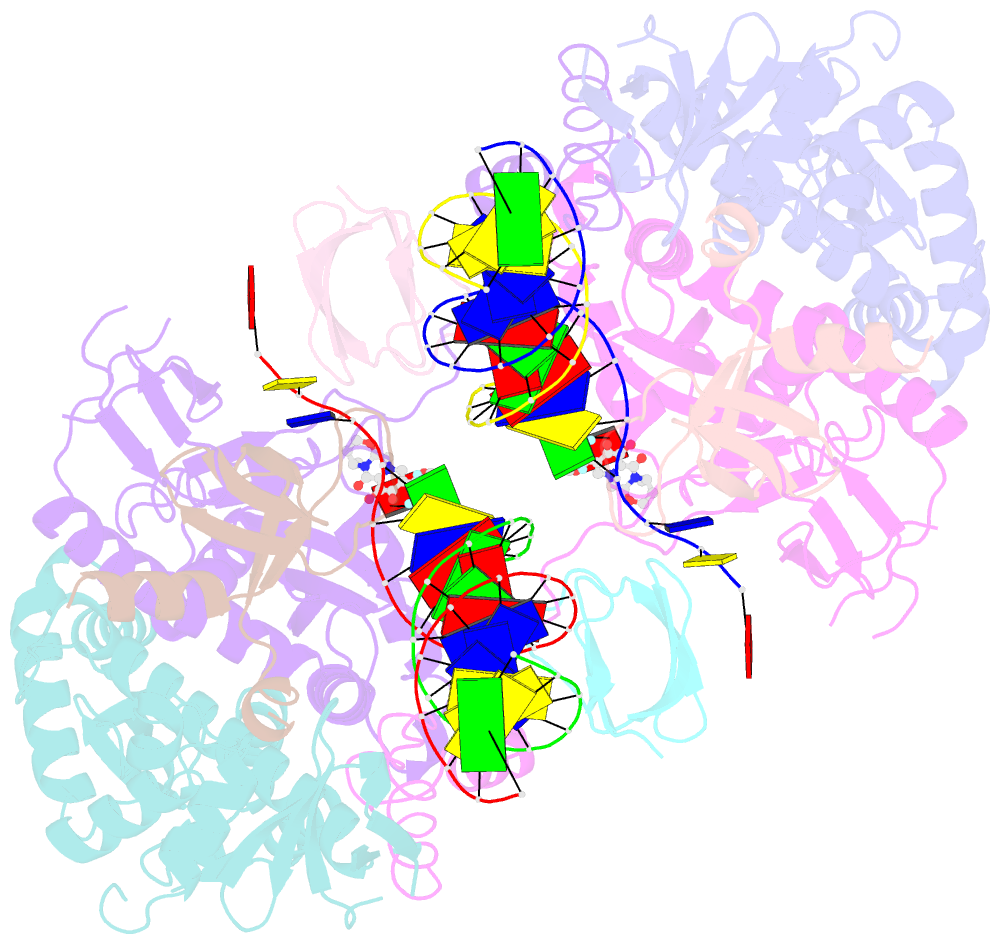
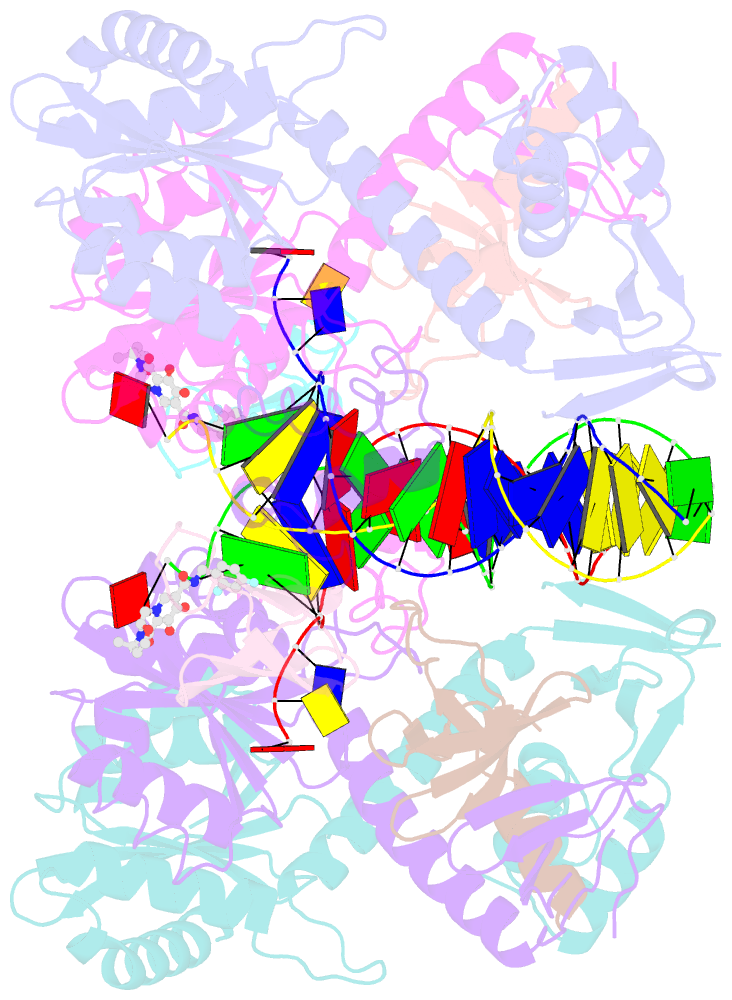
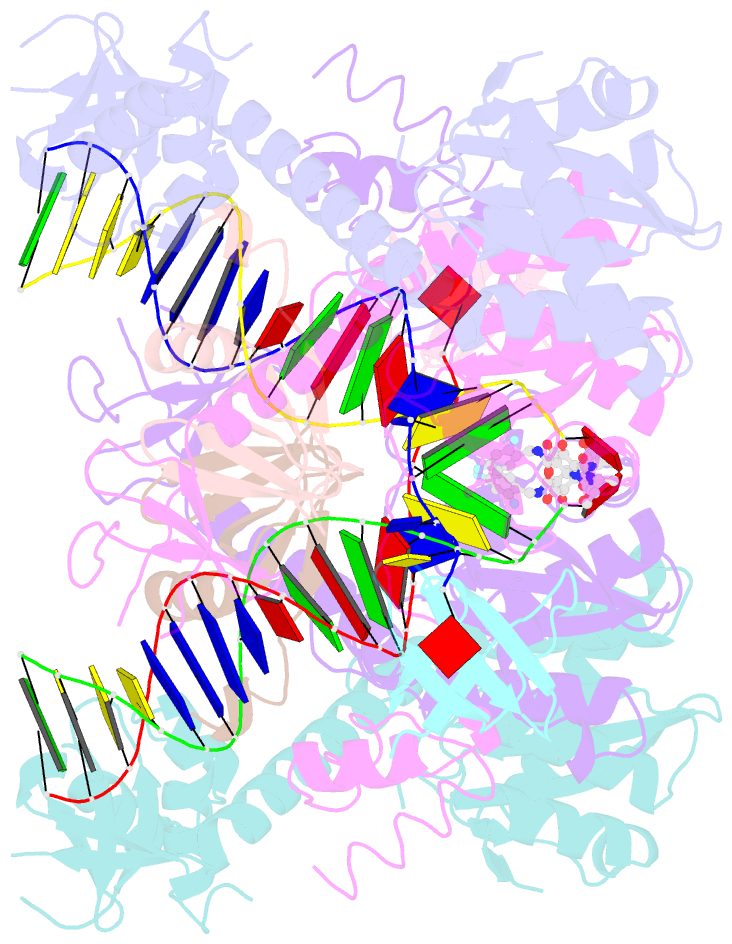
- Structure of e138k hiv-1 intasome with dolutegravir bound
- Reference
- Li M, Oliveira Passos D, Shan Z, Smith SJ, Sun Q, Biswas A, Choudhuri I, Strutzenberg TS, Haldane A, Deng N, Li Z, Zhao XZ, Briganti L, Kvaratskhelia M, Burke Jr TR, Levy RM, Hughes SH, Craigie R, Lyumkis D (2023): "Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants." Sci Adv, 9, eadg5953. doi: 10.1126/sciadv.adg5953.
- Abstract
- HIV-1 infection depends on the integration of viral DNA into host chromatin. Integration is mediated by the viral enzyme integrase and is blocked by integrase strand transfer inhibitors (INSTIs), first-line antiretroviral therapeutics widely used in the clinic. Resistance to even the best INSTIs is a problem, and the mechanisms of resistance are poorly understood. Here, we analyze combinations of the mutations E138K, G140A/S, and Q148H/K/R, which confer resistance to INSTIs. The investigational drug 4d more effectively inhibited the mutants compared with the approved drug Dolutegravir (DTG). We present 11 new cryo-EM structures of drug-resistant HIV-1 intasomes bound to DTG or 4d, with better than 3-Å resolution. These structures, complemented with free energy simulations, virology, and enzymology, explain the mechanisms of DTG resistance involving E138K + G140A/S + Q148H/K/R and show why 4d maintains potency better than DTG. These data establish a foundation for further development of INSTIs that potently inhibit resistant forms in integrase.