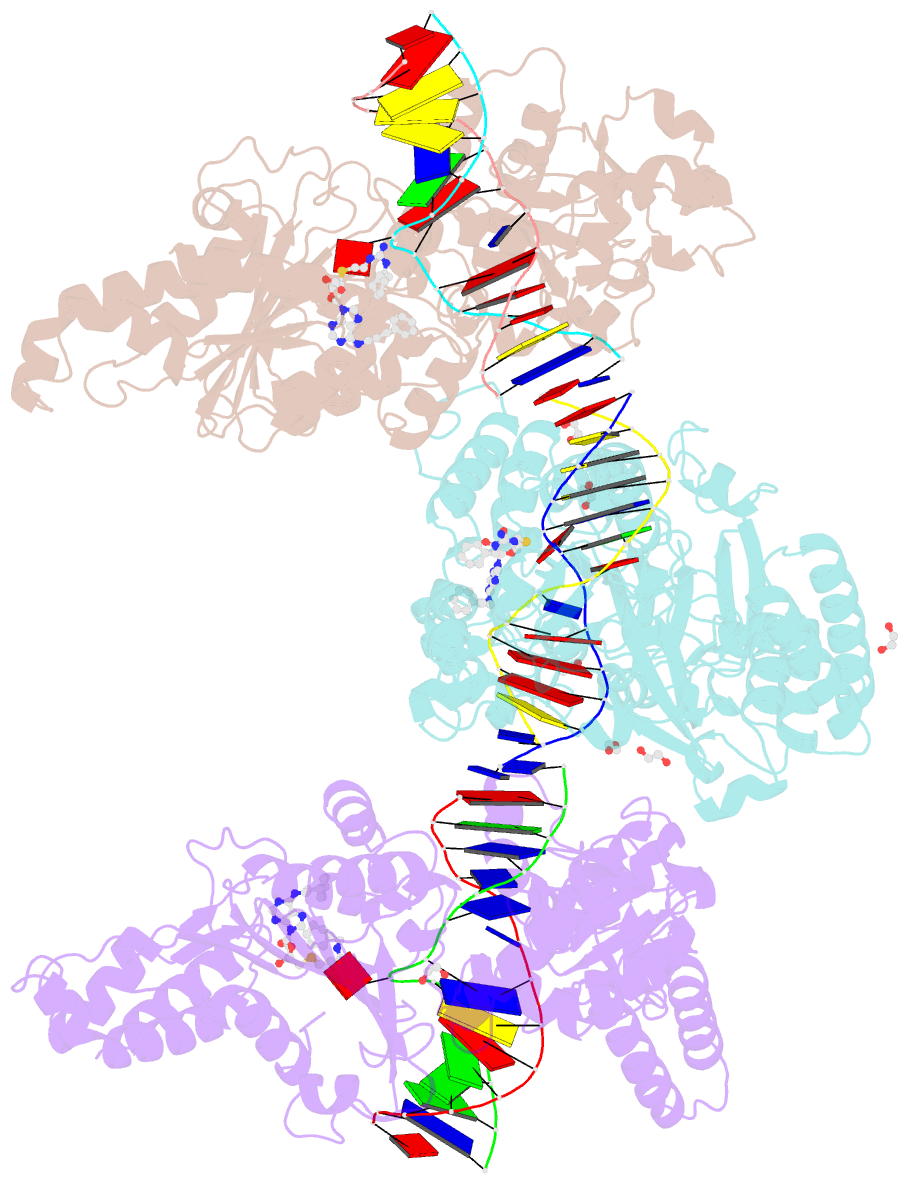
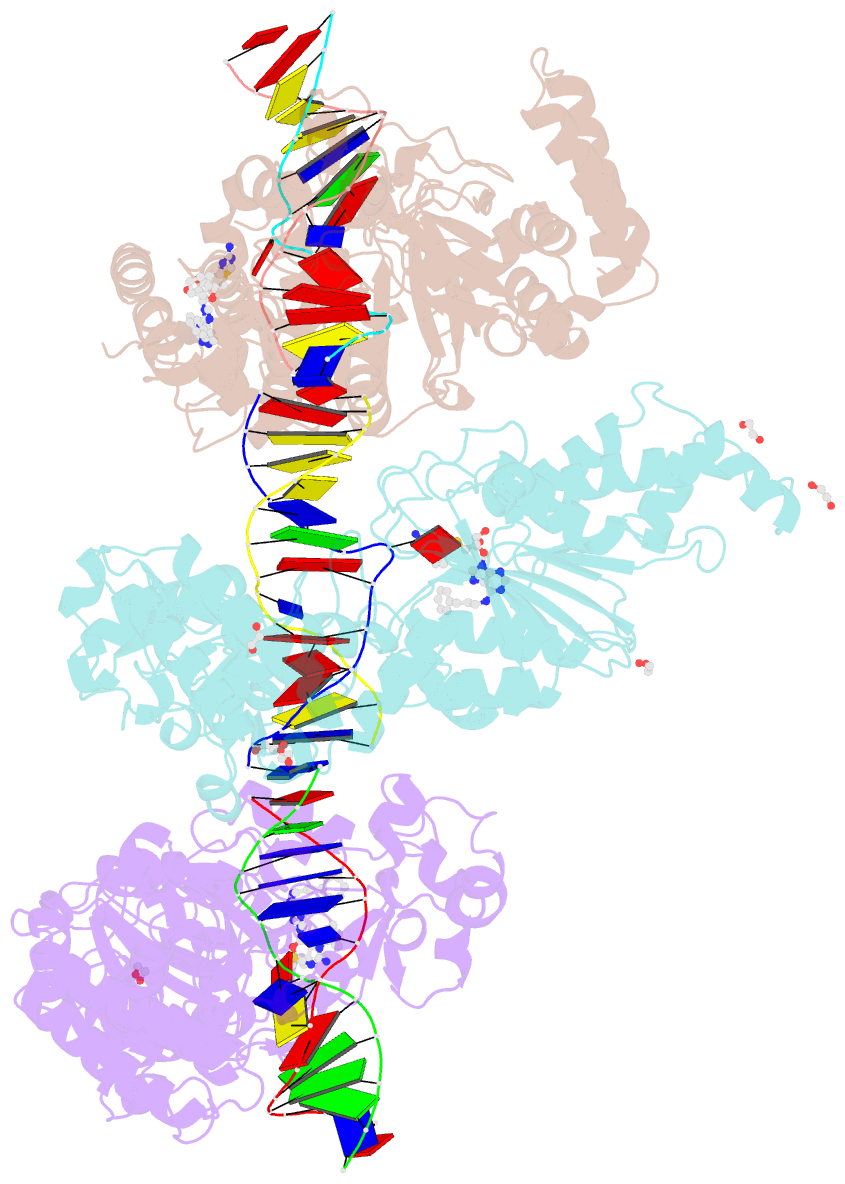
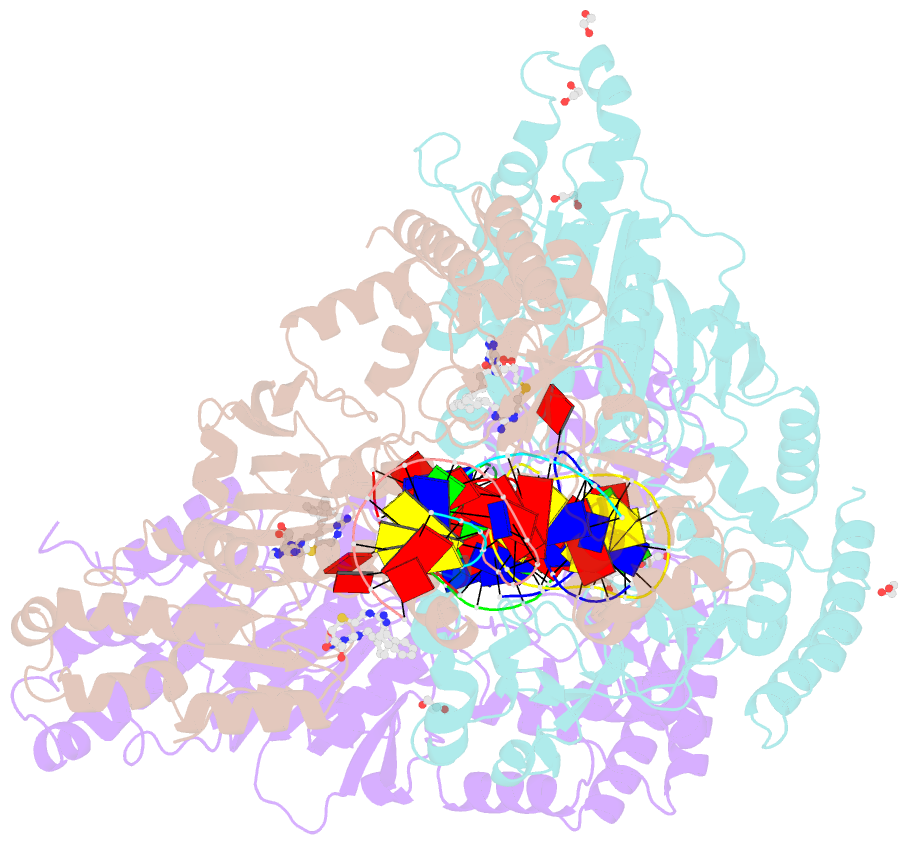
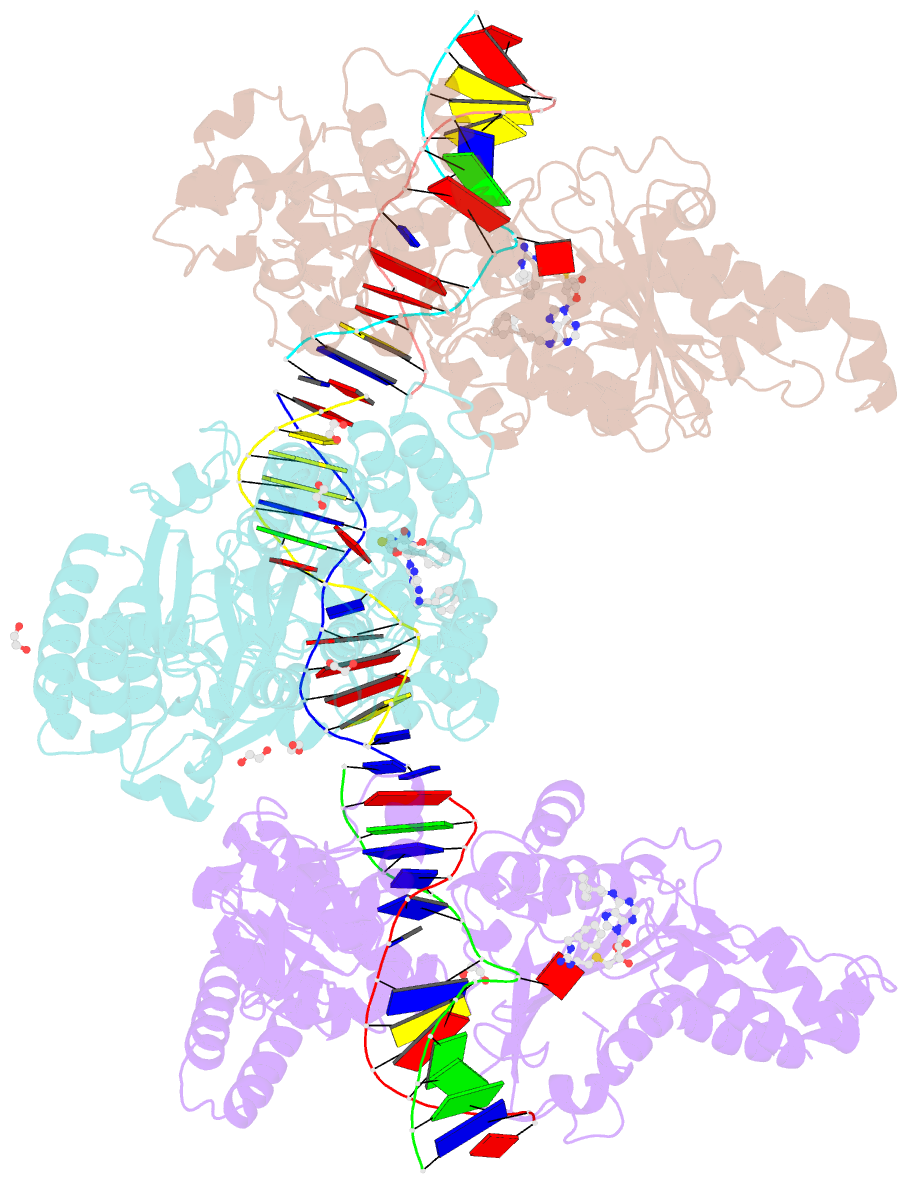
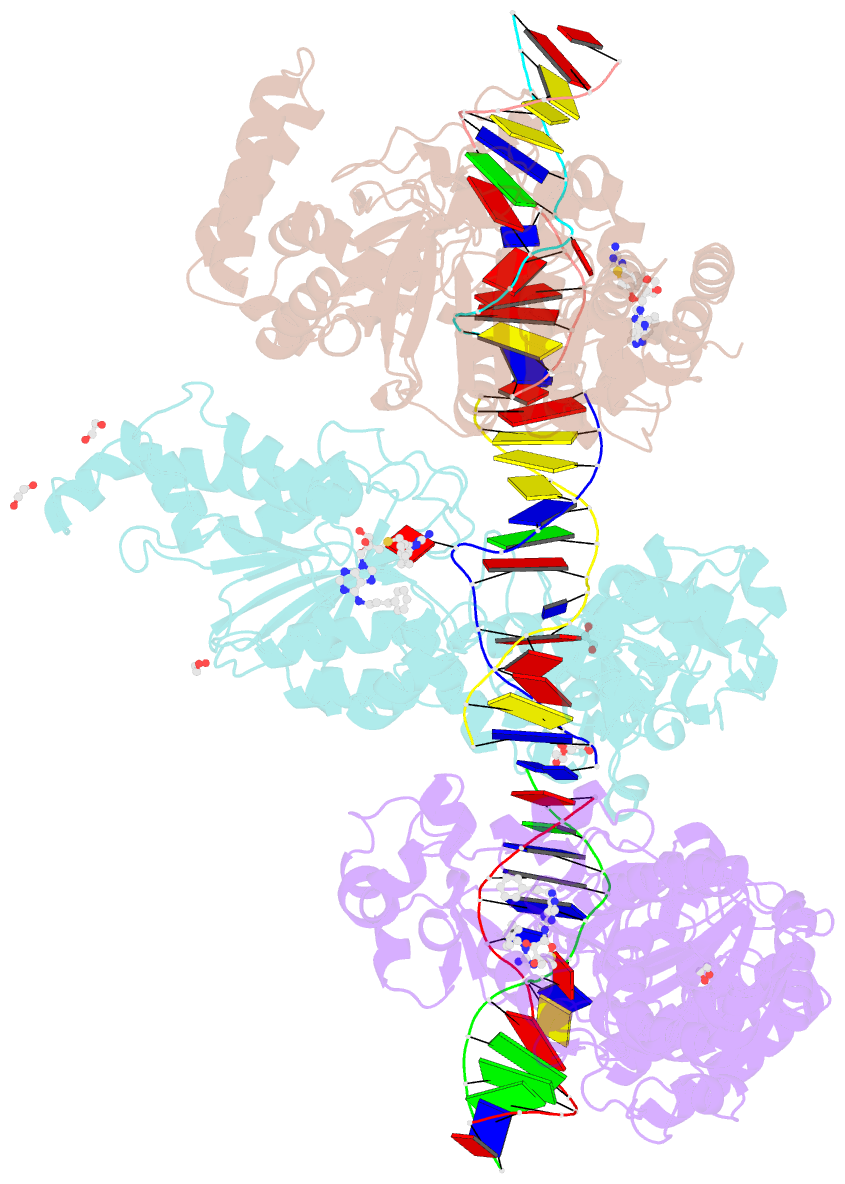
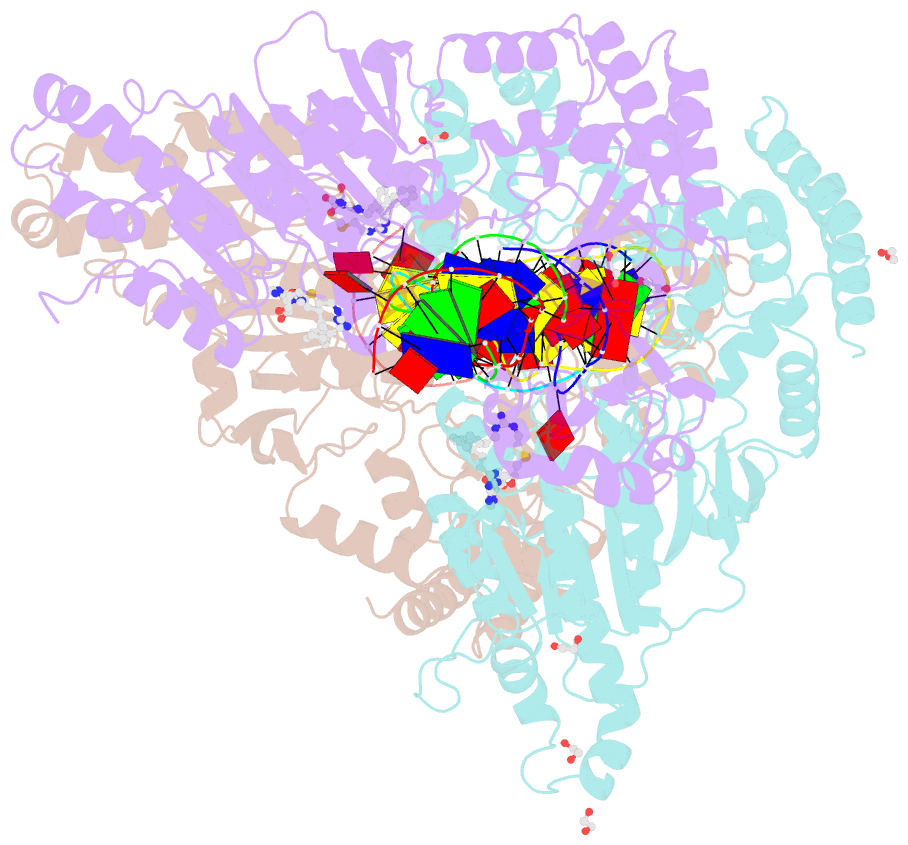
Summary information and primary citation
- PDB-id
- 8fs1; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA-inhibitor
- Method
- X-ray (2.74 Å)
- Summary
- Cama adenine methyltransferase complexed to cognate substrate DNA and inhibitor 11a (yd905)
- Reference
- Zhou J, Deng Y, Iyamu ID, Horton JR, Yu D, Hajian T, Vedadi M, Rotili D, Mai A, Blumenthal RM, Zhang X, Huang R, Cheng X (2023): "Comparative Study of Adenosine Analogs as Inhibitors of Protein Arginine Methyltransferases and a Clostridioides difficile- Specific DNA Adenine Methyltransferase." Acs Chem.Biol., 18, 734-745. doi: 10.1021/acschembio.3c00035.
- Abstract
- S-Adenosyl-l-methionine (SAM) analogs are adaptable tools for studying and therapeutically inhibiting SAM-dependent methyltransferases (MTases). Some MTases play significant roles in host-pathogen interactions, one of which is Clostridioides difficile-specific DNA adenine MTase (CamA). CamA is needed for efficient sporulation and alters persistence in the colon. To discover potent and selective CamA inhibitors, we explored modifications of the solvent-exposed edge of the SAM adenosine moiety. Starting from the two parental compounds (