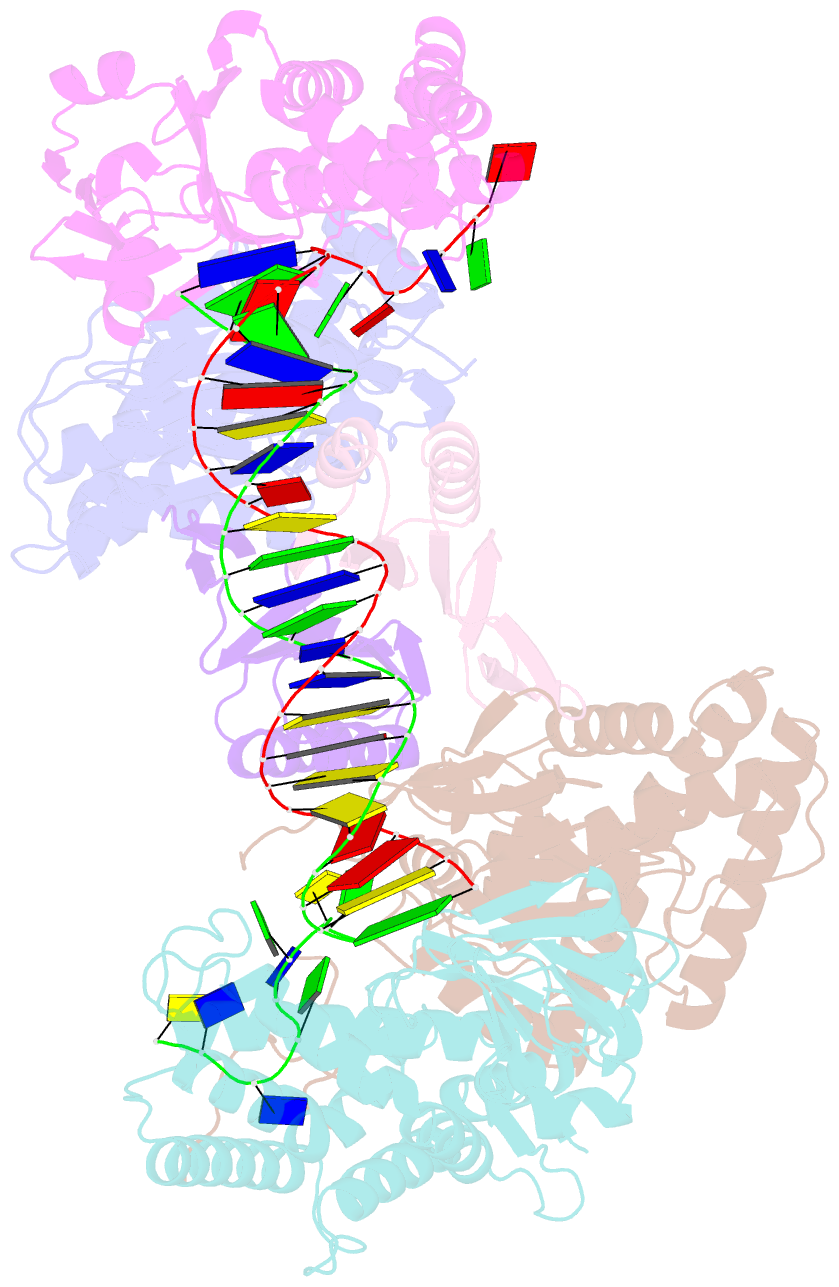
Summary information and primary citation
- PDB-id
- 8fya; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- cryo-EM (2.91 Å)
- Summary
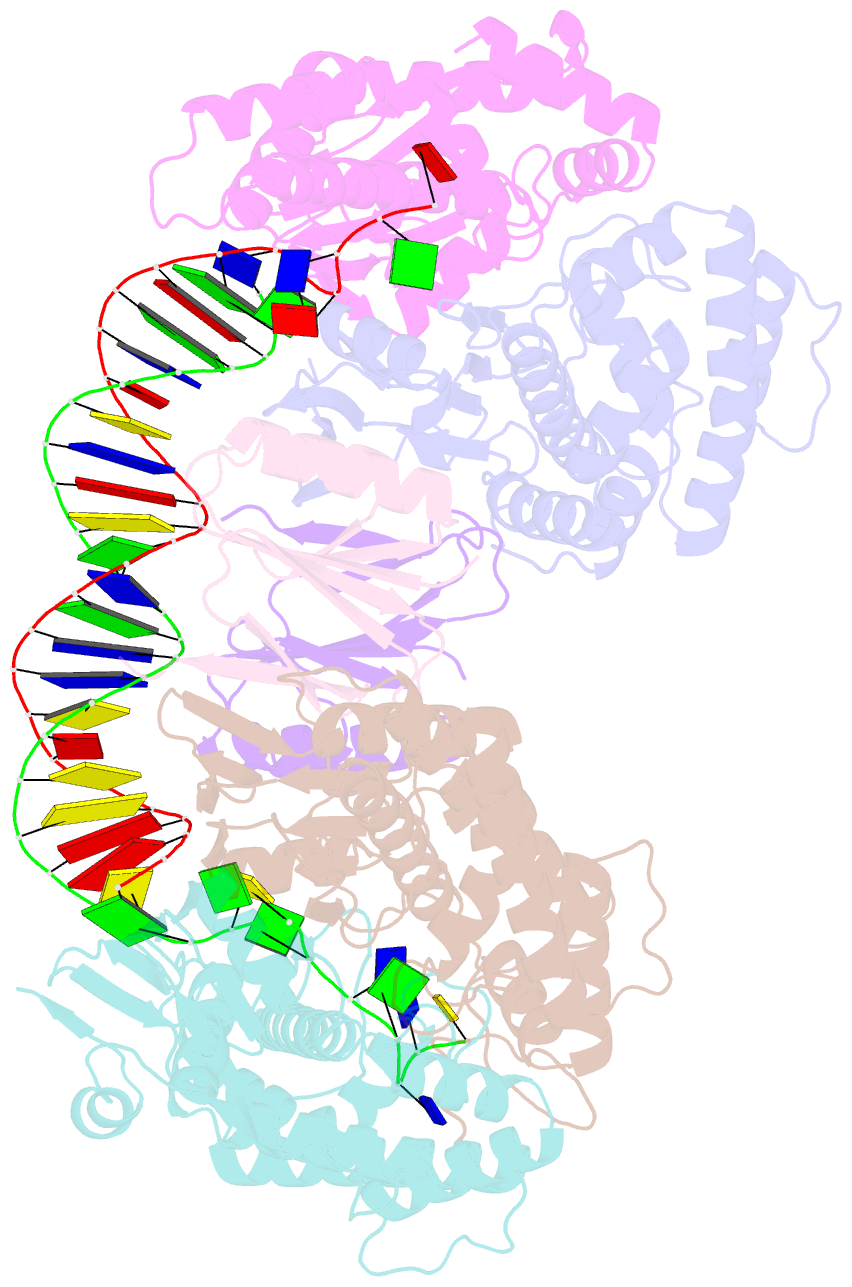
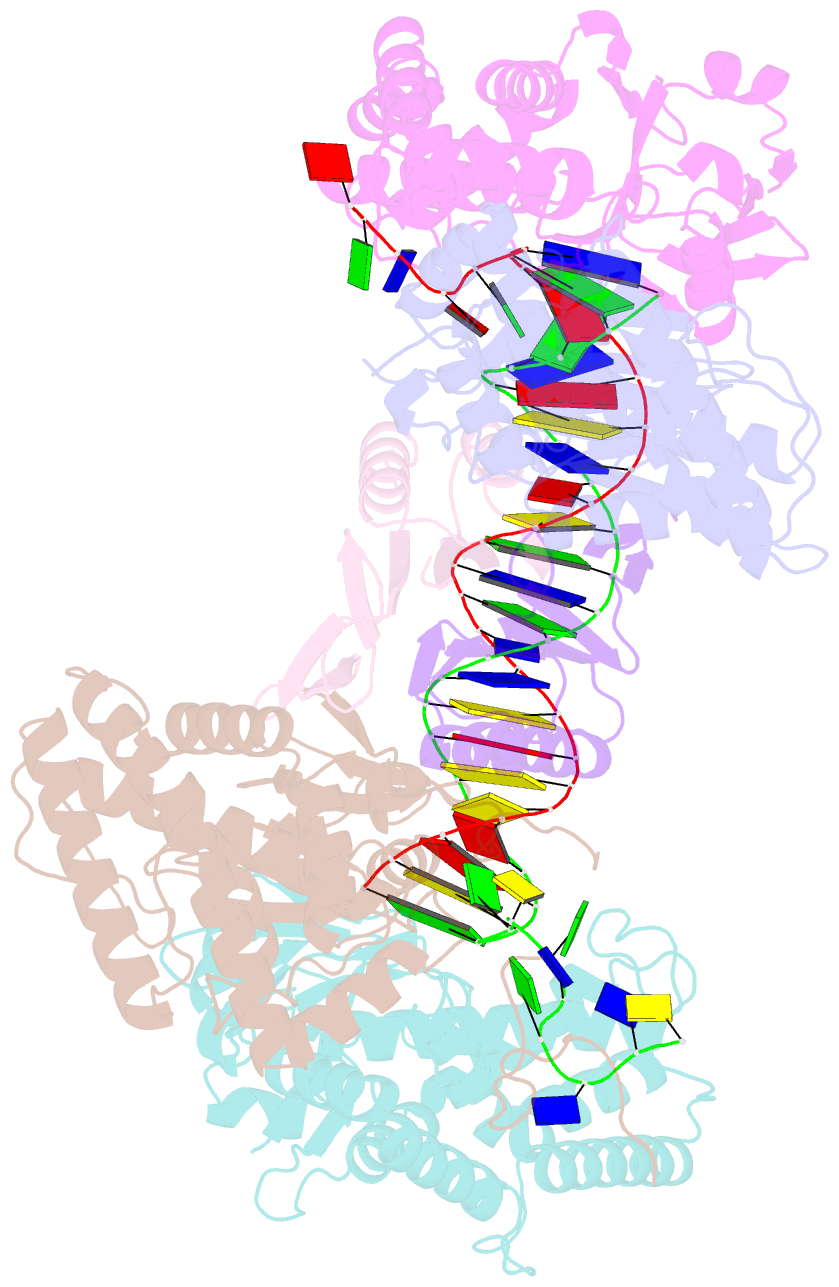
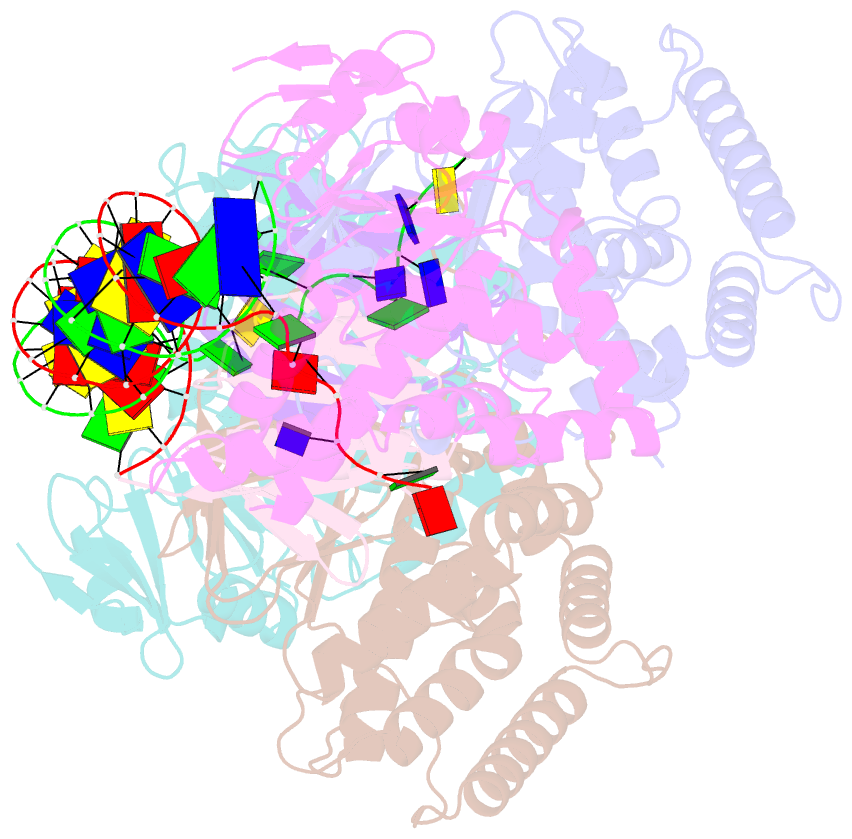
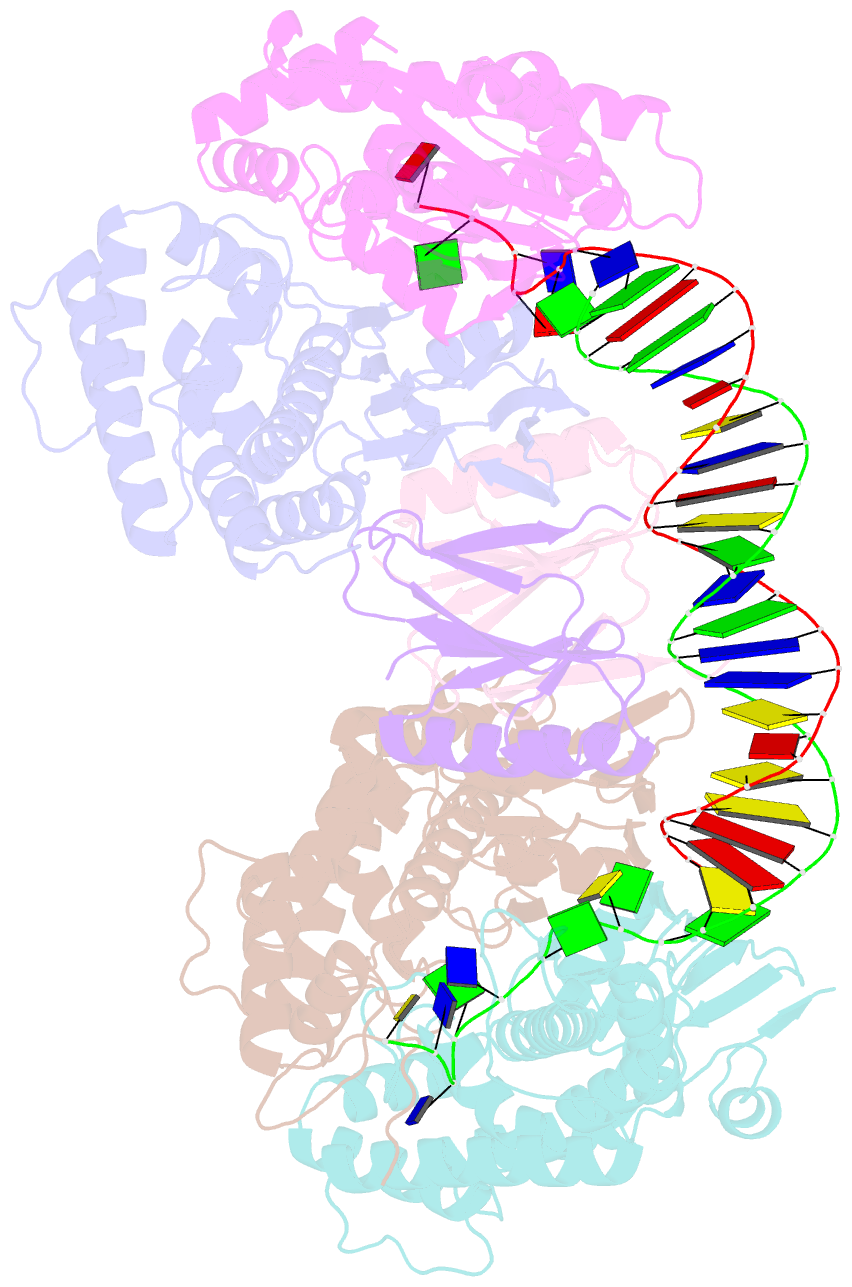
- cryo-EM structure of cas1:cas2-deddh:pam-containing prespacer complex
- Reference
- Wang JY, Tuck OT, Skopintsev P, Soczek KM, Li G, Al-Shayeb B, Zhou J, Doudna JA (2023): "Genome expansion by a CRISPR trimmer-integrase." Nature, 618, 855-861. doi: 10.1038/s41586-023-06178-2.
- Abstract
- CRISPR-Cas adaptive immune systems capture DNA fragments from invading mobile genetic elements and integrate them into the host genome to provide a template for RNA-guided immunity1. CRISPR systems maintain genome integrity and avoid autoimmunity by distinguishing between self and non-self, a process for which the CRISPR/Cas1-Cas2 integrase is necessary but not sufficient2-5. In some microorganisms, the Cas4 endonuclease assists CRISPR adaptation6,7, but many CRISPR-Cas systems lack Cas48. Here we show here that an elegant alternative pathway in a type I-E system uses an internal DnaQ-like exonuclease (DEDDh) to select and process DNA for integration using the protospacer adjacent motif (PAM). The natural Cas1-Cas2/exonuclease fusion (trimmer-integrase) catalyses coordinated DNA capture, trimming and integration. Five cryo-electron microscopy structures of the CRISPR trimmer-integrase, visualized both before and during DNA integration, show how asymmetric processing generates size-defined, PAM-containing substrates. Before genome integration, the PAM sequence is released by Cas1 and cleaved by the exonuclease, marking inserted DNA as self and preventing aberrant CRISPR targeting of the host. Together, these data support a model in which CRISPR systems lacking Cas4 use fused or recruited9,10 exonucleases for faithful acquisition of new CRISPR immune sequences.