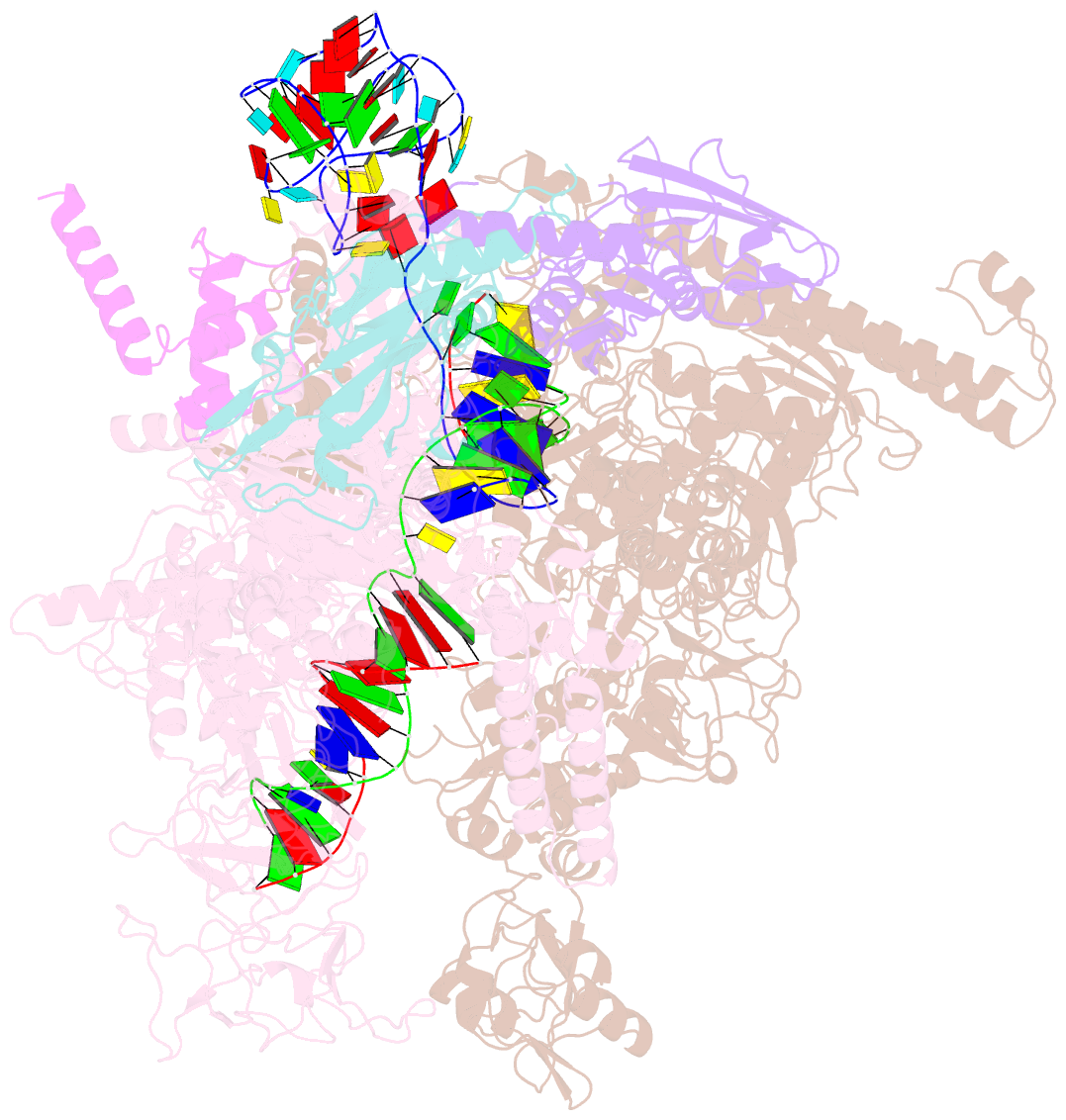
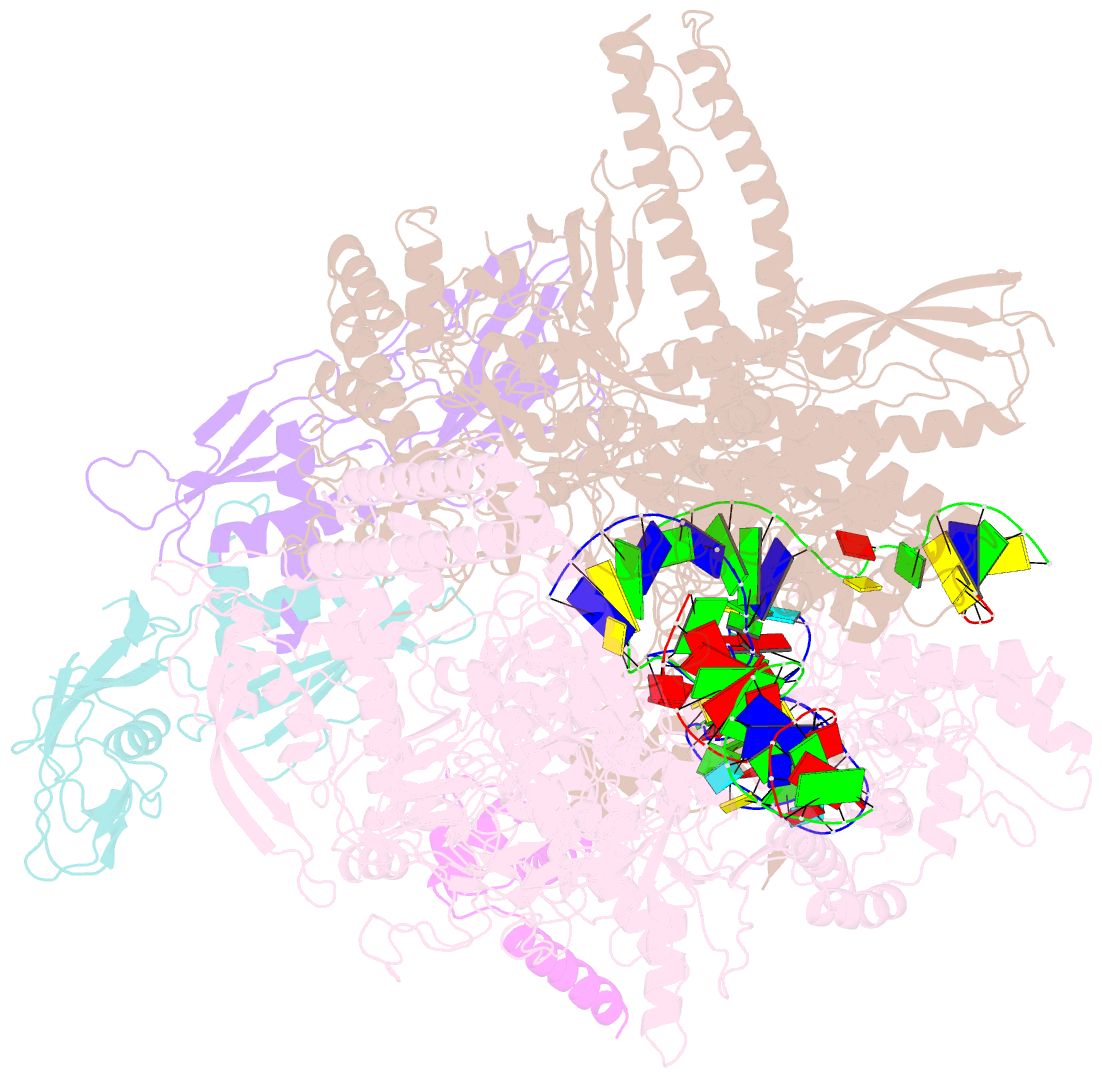
Summary information and primary citation
- PDB-id
- 8g00; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- cryo-EM (3.4 Å)
- Summary
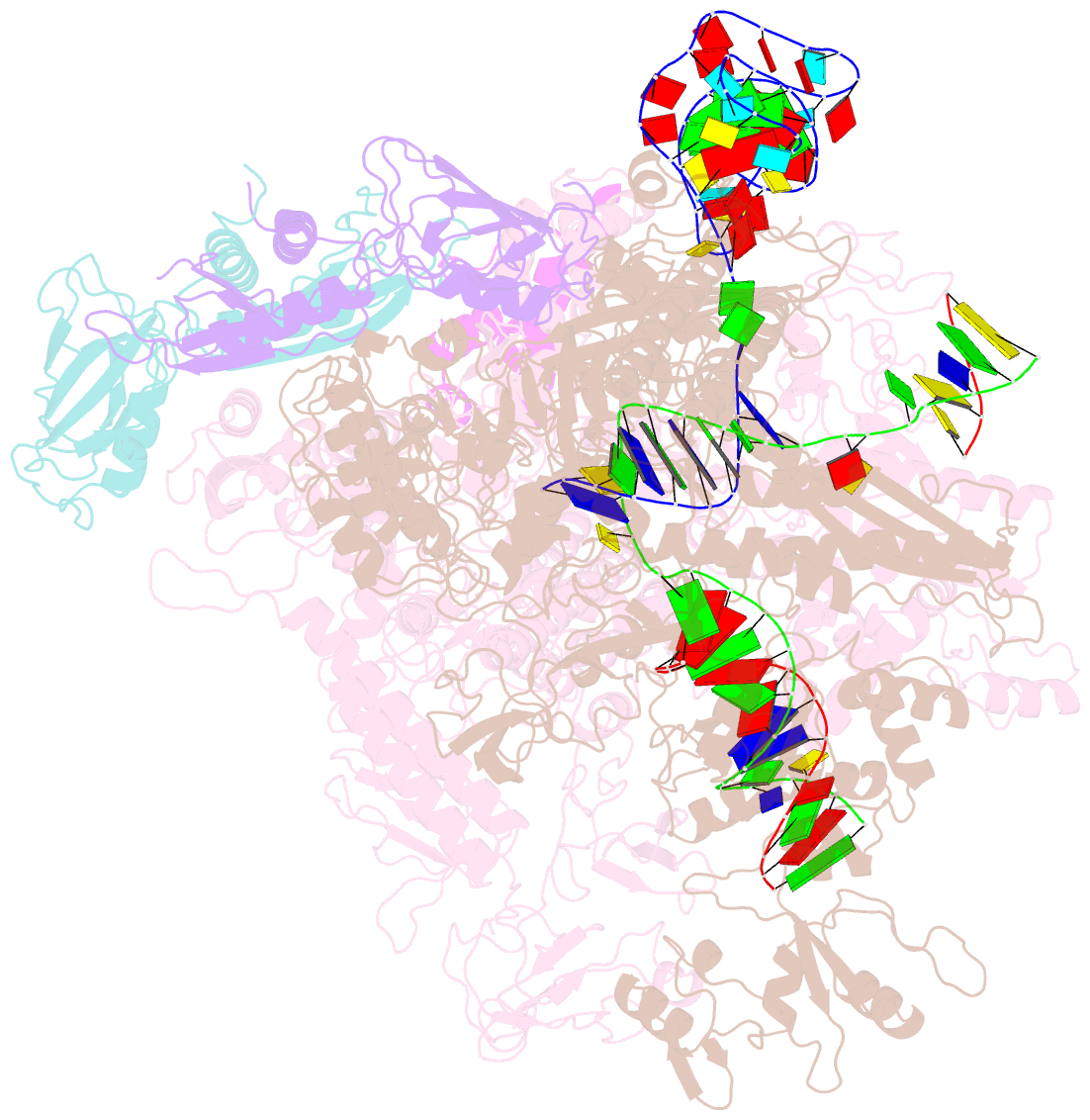
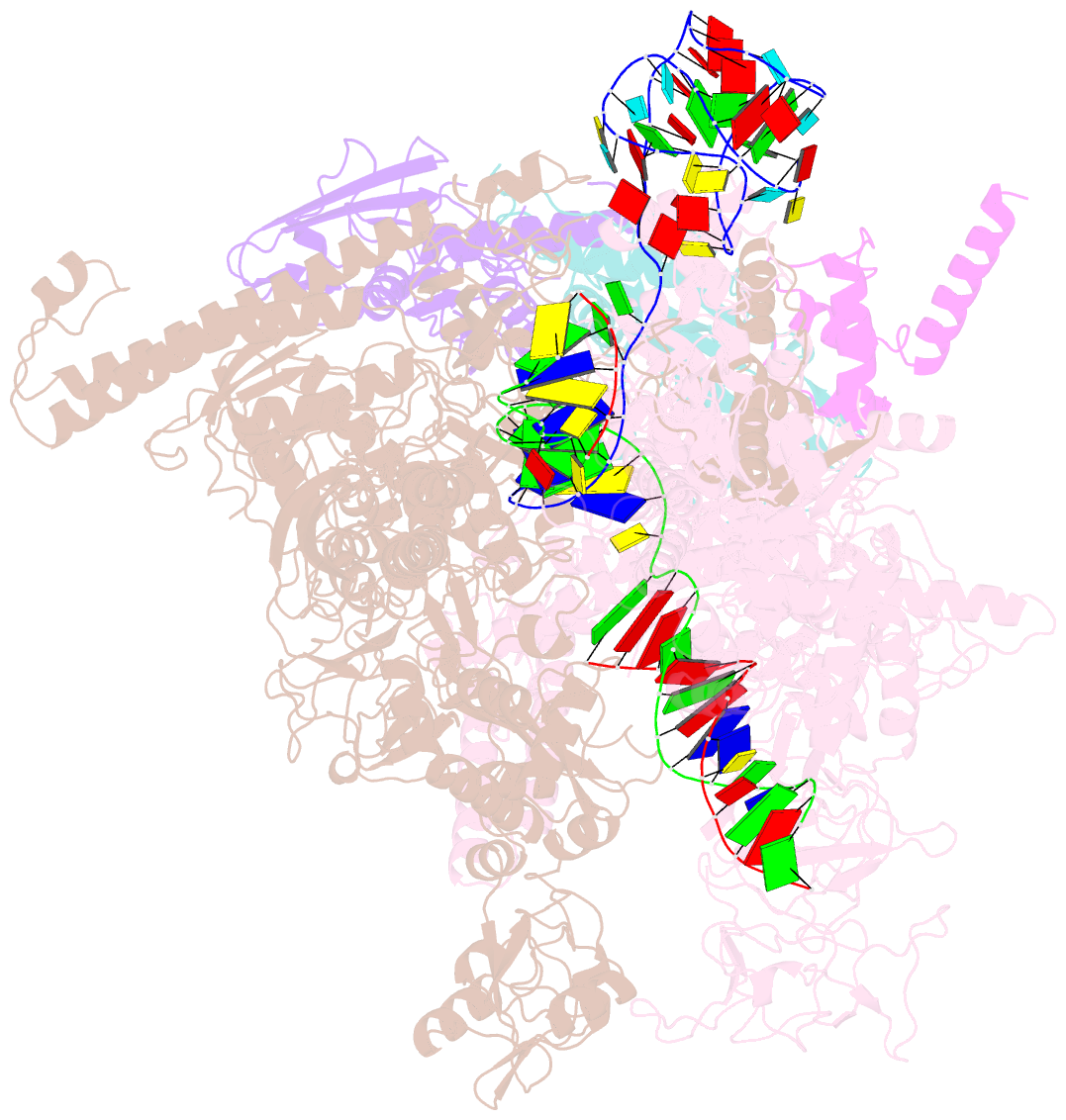
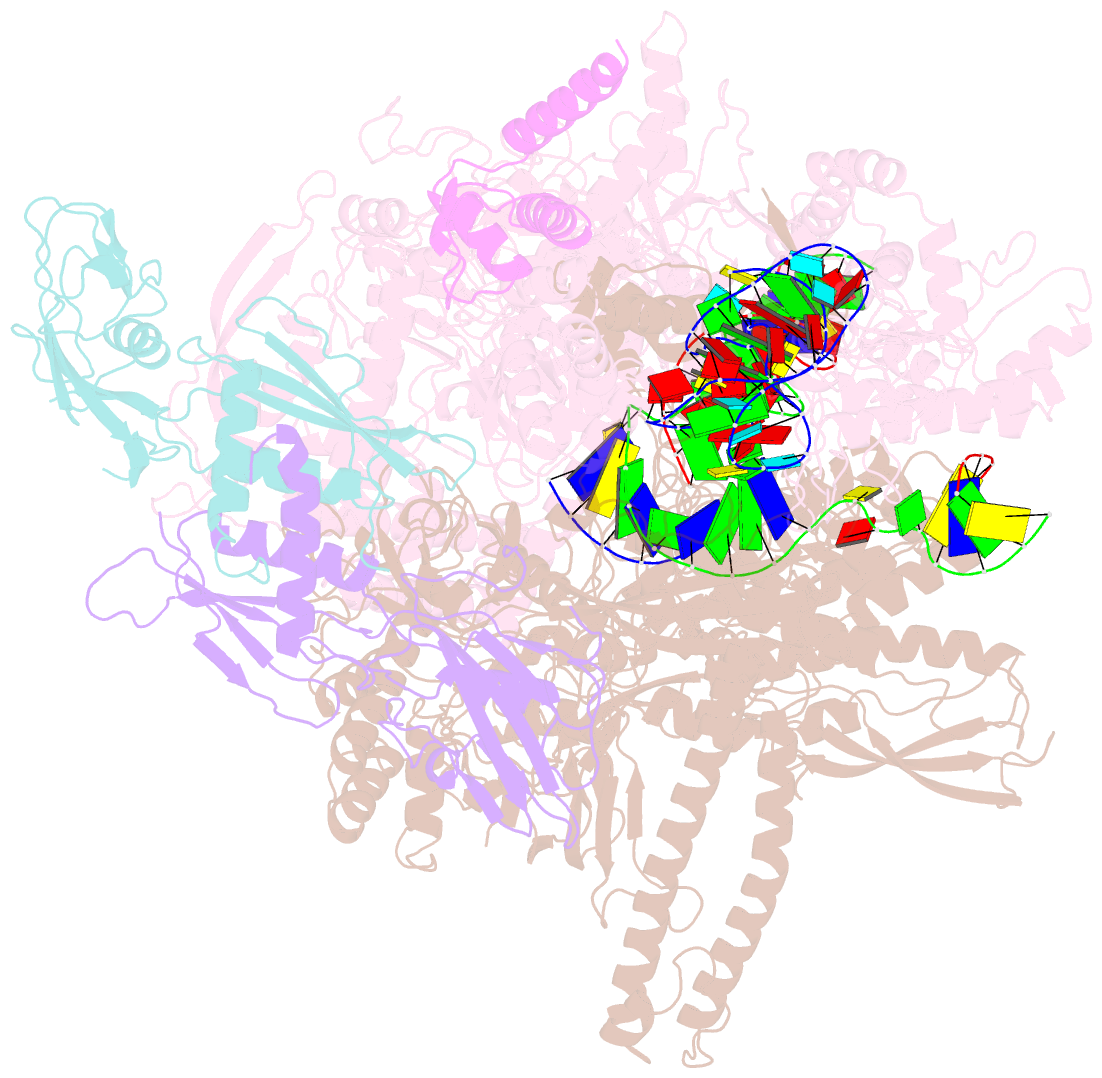
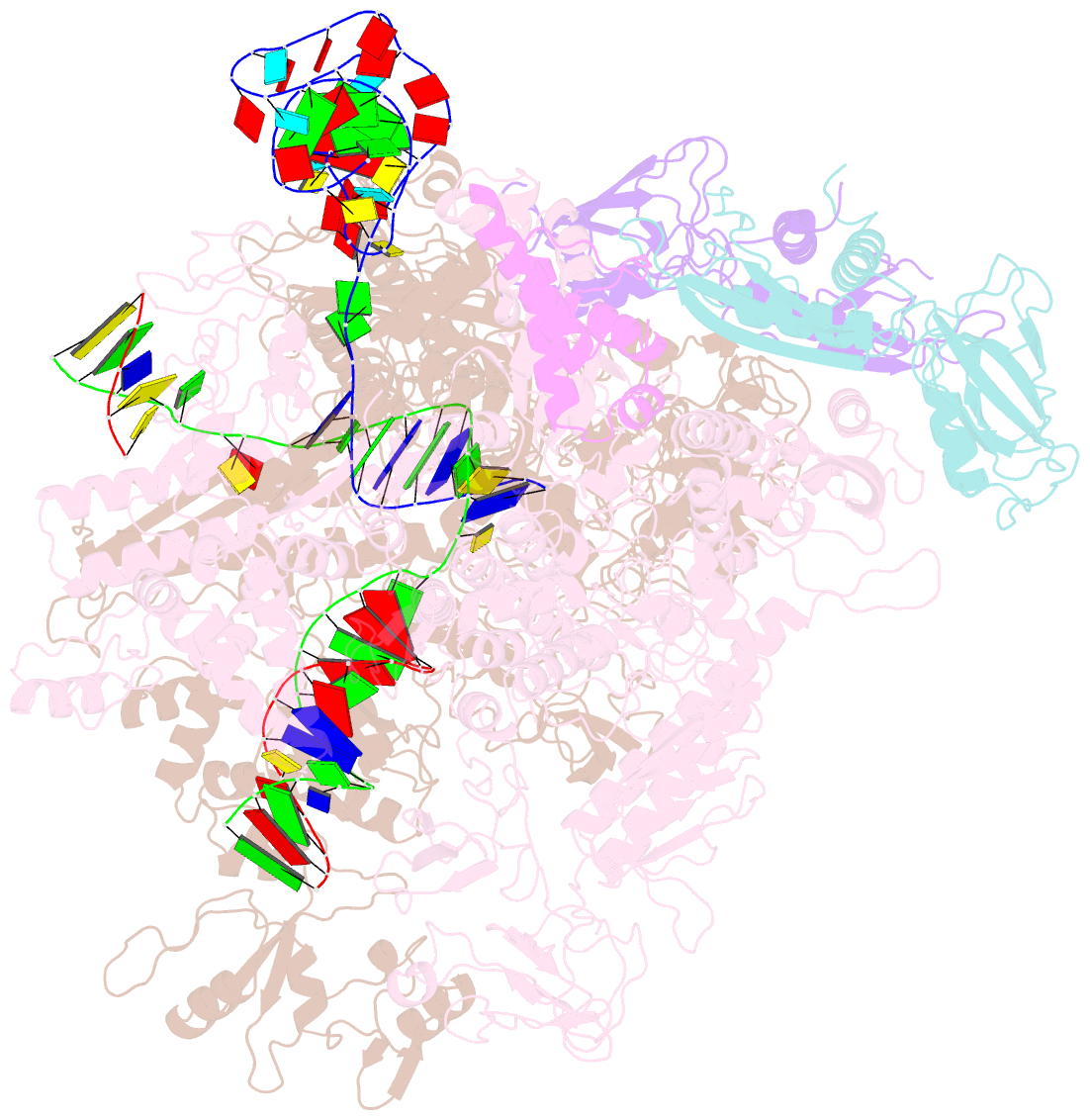
- cryo-EM structure of 3dva component 0 of escherichia coli que-pec (paused elongation complex) RNA polymerase minus preq1 ligand
- Reference
- Chauvier A, Porta JC, Deb I, Ellinger E, Meze K, Frank AT, Ohi MD, Walter NG (2023): "Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand." Nat.Struct.Mol.Biol., 30, 902-913. doi: 10.1038/s41594-023-01002-x.
- Abstract
- Folding of nascent transcripts can be modulated by the RNA polymerase (RNAP) that carries out their transcription, and vice versa. A pause of RNAP during transcription of a preQ1 riboswitch (termed que-PEC) is stabilized by a previously characterized template consensus sequence and the ligand-free conformation of the nascent RNA. Ligand binding to the riboswitch induces RNAP pause release and downstream transcription termination; however, the mechanism by which riboswitch folding modulates pausing is unclear. Here, we report single-particle cryo-electron microscopy reconstructions of que-PEC in ligand-free and ligand-bound states. In the absence of preQ1, the RNA transcript is in an unexpected hyper-translocated state, preventing downstream nucleotide incorporation. Strikingly, on ligand binding, the riboswitch rotates around its helical axis, expanding the surrounding RNAP exit channel and repositioning the transcript for elongation. Our study reveals the tight coupling by which nascent RNA structures and their ligands can functionally regulate the macromolecular transcription machinery.