Summary information and primary citation
- PDB-id
- 8g0h; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (3.8 Å)
- Summary
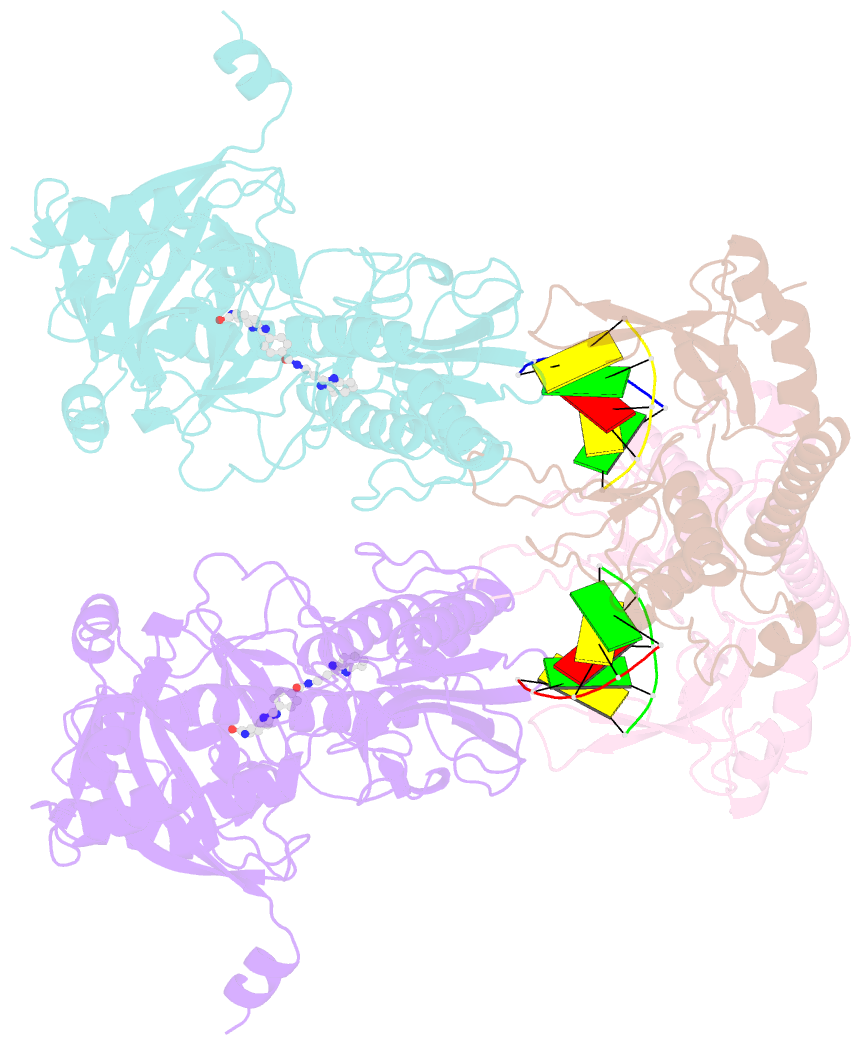
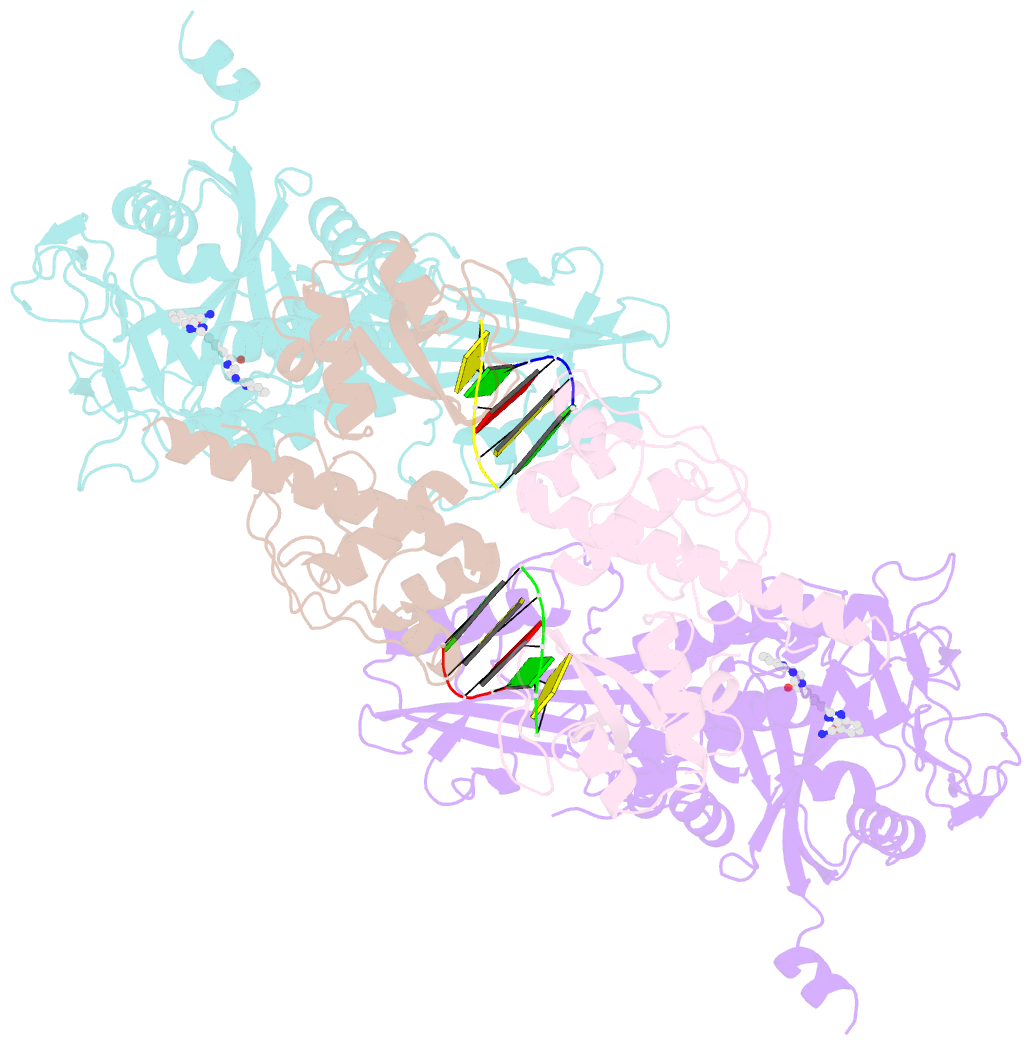
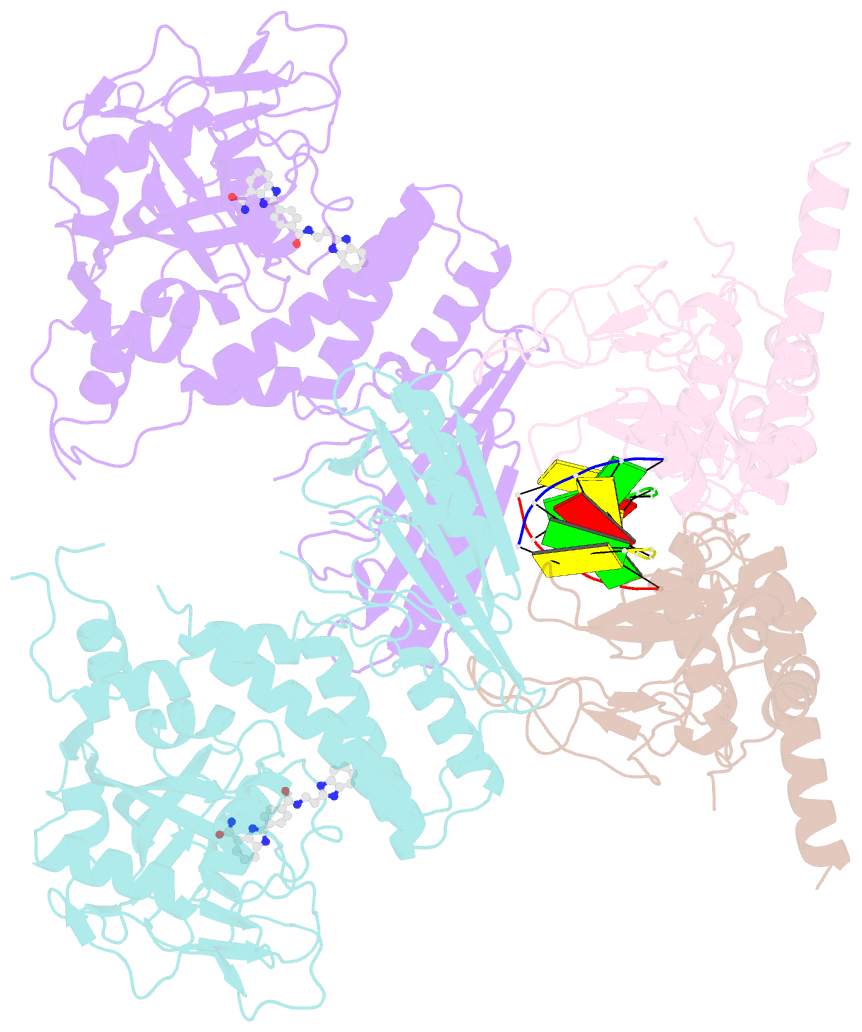
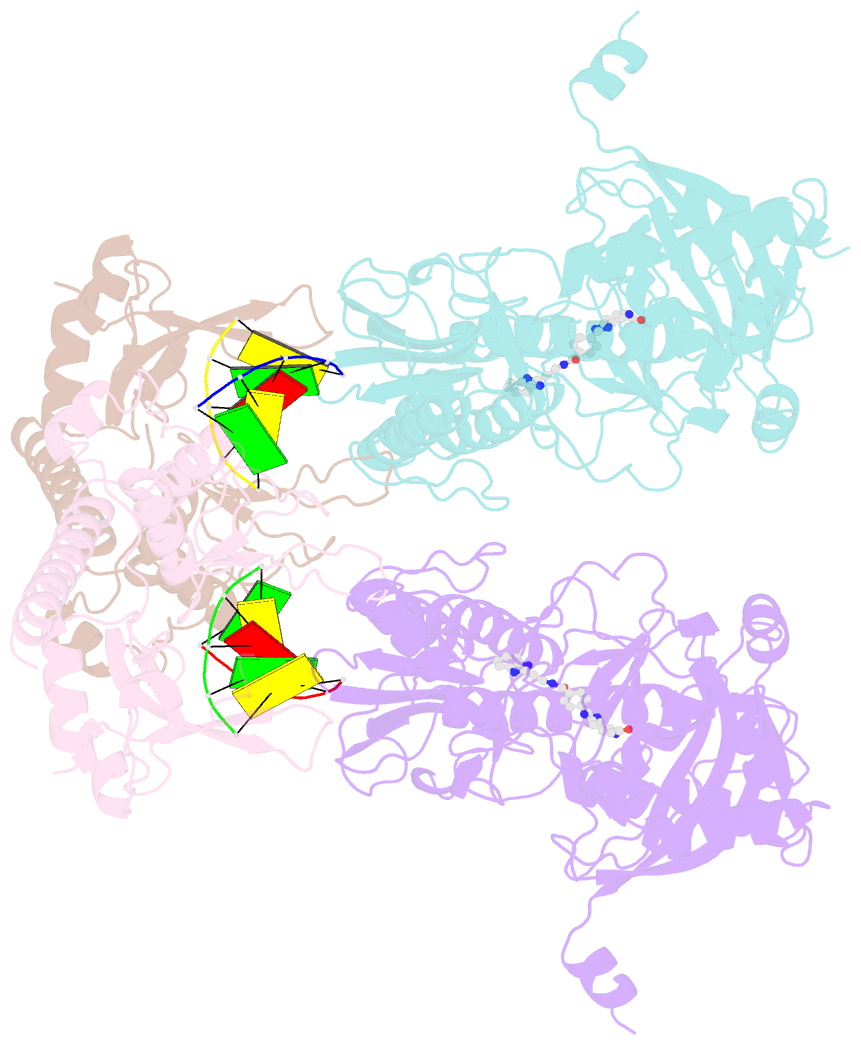
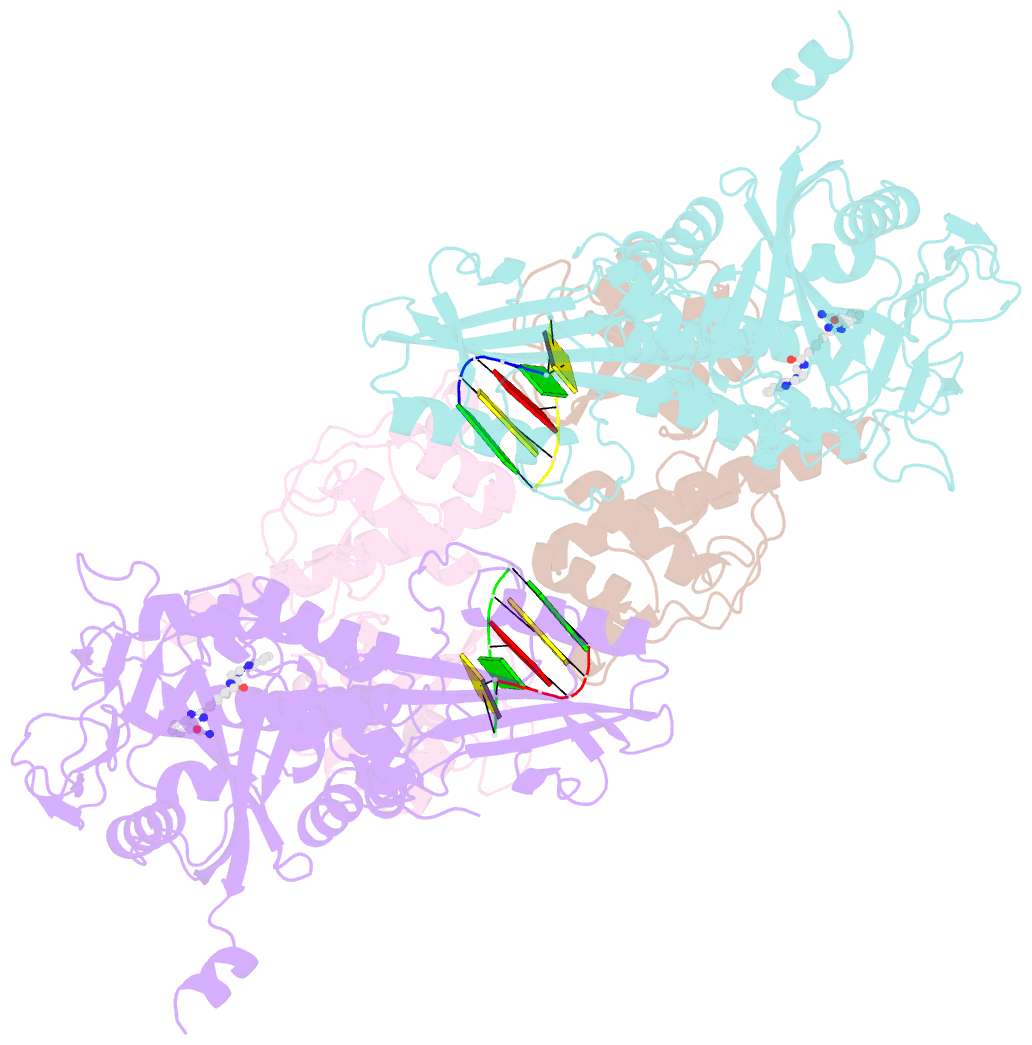
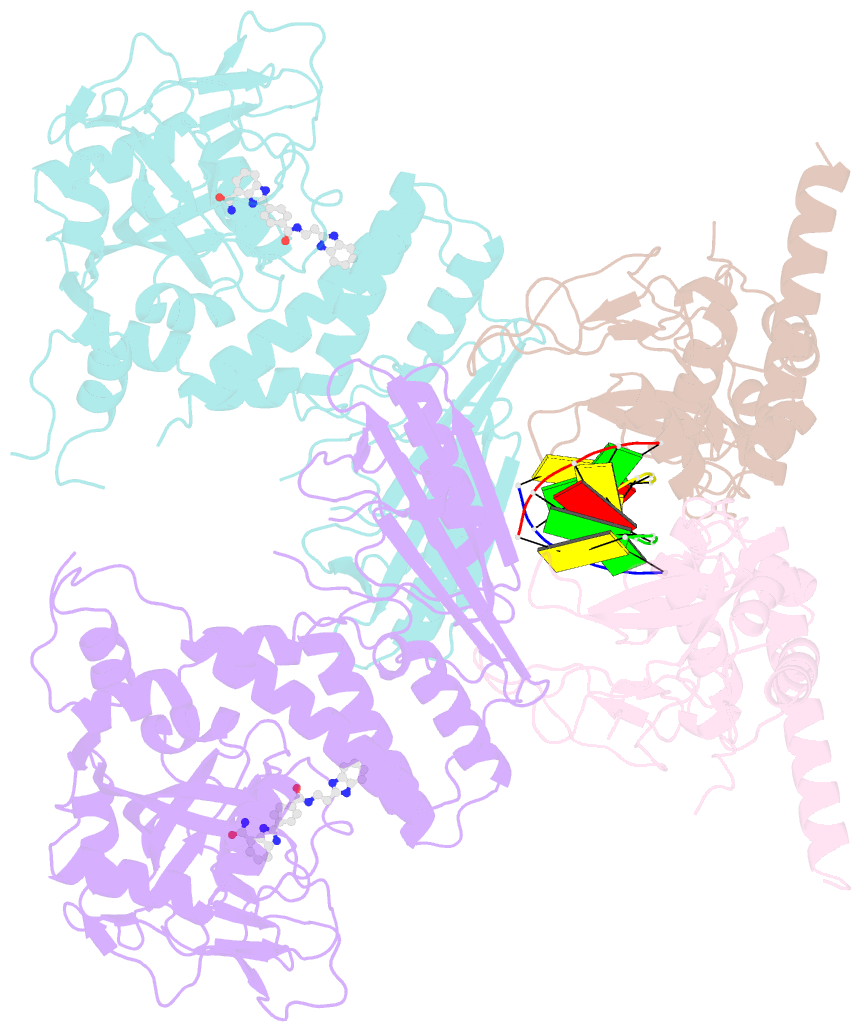
- Human parp1 deltav687-e688 bound to uktt5 (compound 10) and to a DNA double strand break.
- Reference
- Velagapudi UK, Rouleau-Turcotte E, Billur R, Shao X, Patil M, Black BE, Pascal JM, Talele TT (2024): "Novel modifications of PARP inhibitor veliparib increase PARP1 binding to DNA breaks." Biochem.J., 481, 437-460. doi: 10.1042/BCJ20230406.
- Abstract
- Catalytic poly(ADP-ribose) production by PARP1 is allosterically activated through interaction with DNA breaks, and PARP inhibitor compounds have the potential to influence PARP1 allostery in addition to preventing catalytic activity. Using the benzimidazole-4-carboxamide pharmacophore present in the first generation PARP1 inhibitor veliparib, a series of eleven derivatives was designed, synthesized, and evaluated as allosteric PARP1 inhibitors, with the premise that bulky substituents would engage the HD regulatory domain and thereby promote PARP1 retention on DNA breaks. We found that core scaffold modifications could indeed increase PARP1 affinity for DNA; however, the bulk of the modification alone was insufficient to trigger PARP1 allosteric retention on DNA breaks. Rather, compounds eliciting PARP1 retention on DNA breaks were found to be rigidly held in a position that interferes with a specific region of the HD domain, a region that is not targeted by current clinical PARP inhibitors. Collectively, these compounds highlight a unique way to trigger PARP1 retention on DNA breaks and open a path to unveil the pharmacological benefits of such inhibitors with novel properties.