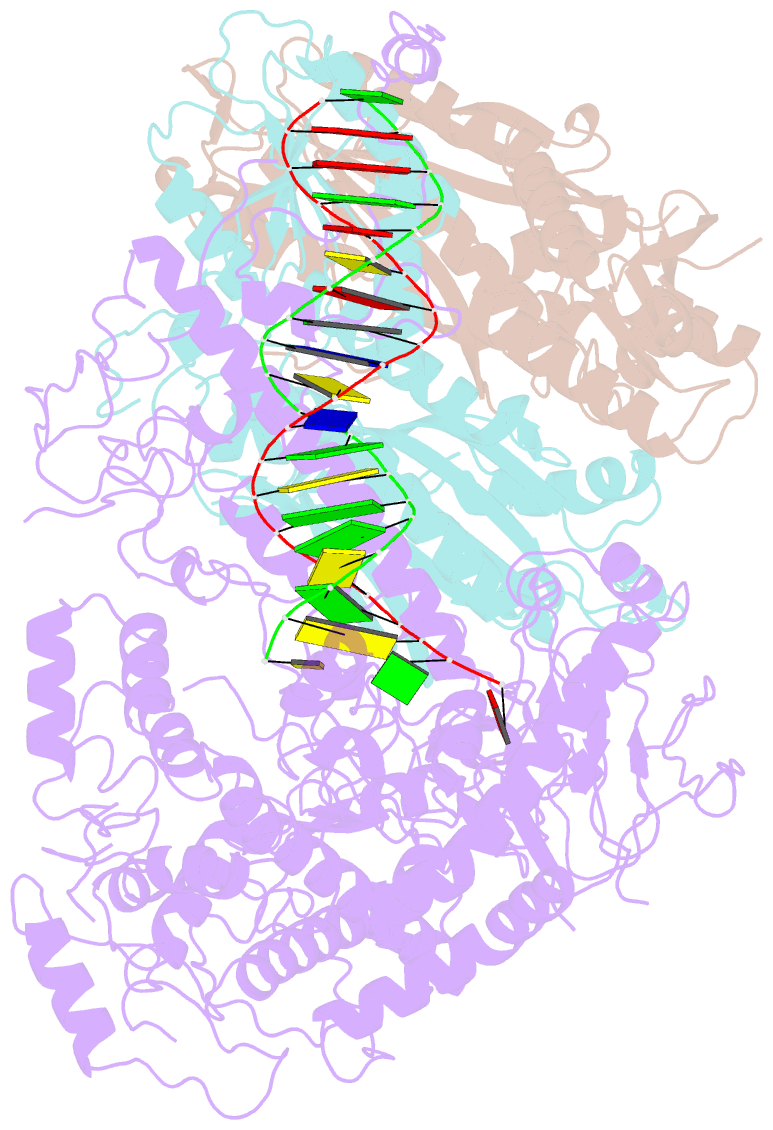
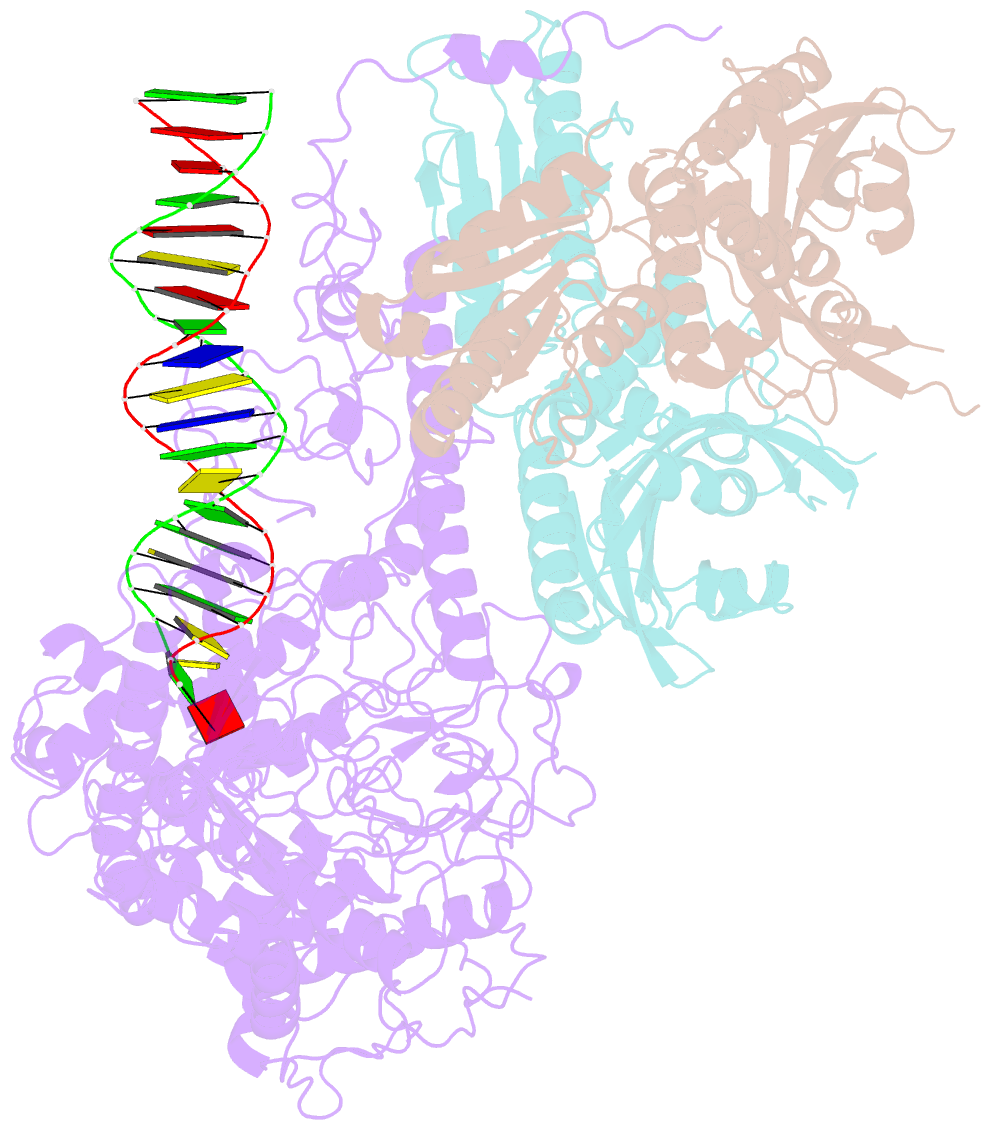
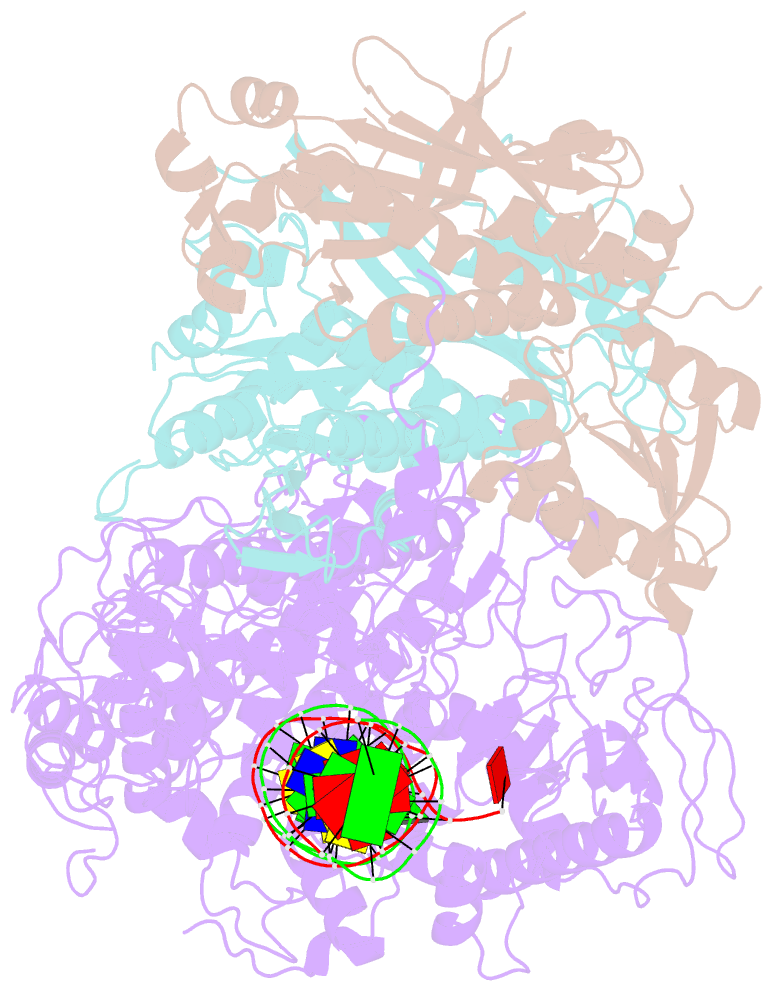
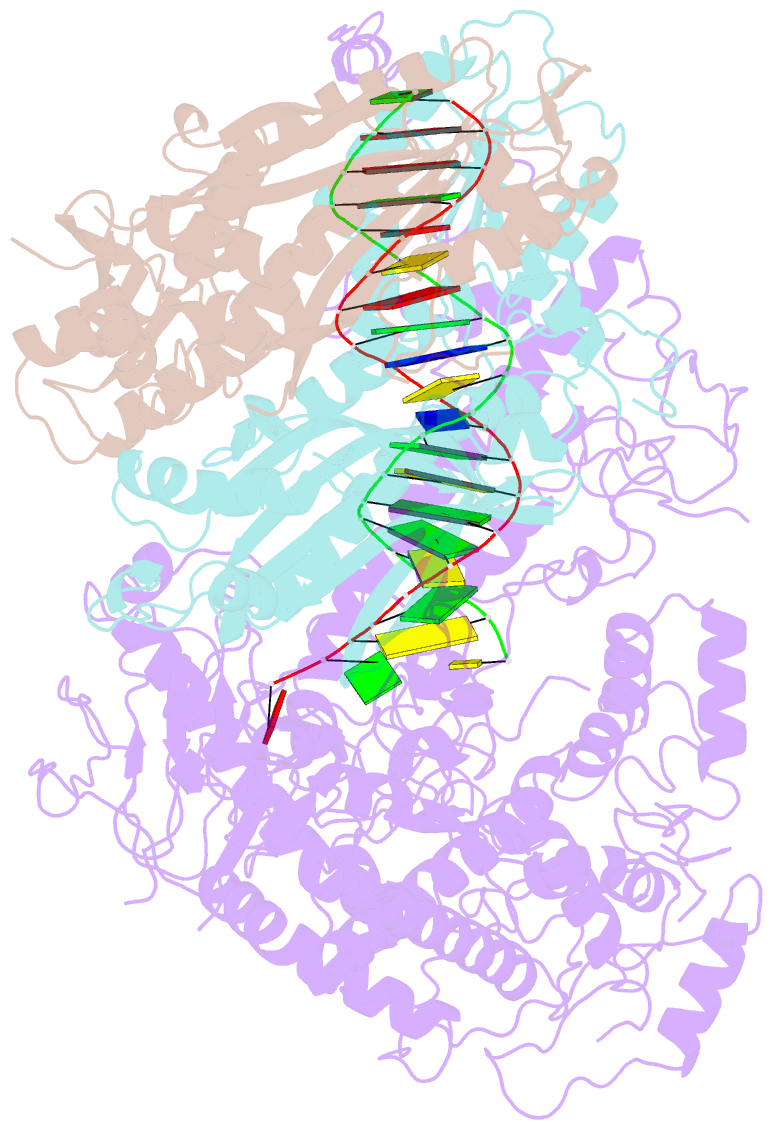
Summary information and primary citation
- PDB-id
- 8g5l; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- replication-DNA
- Method
- cryo-EM (3.0 Å)
- Summary
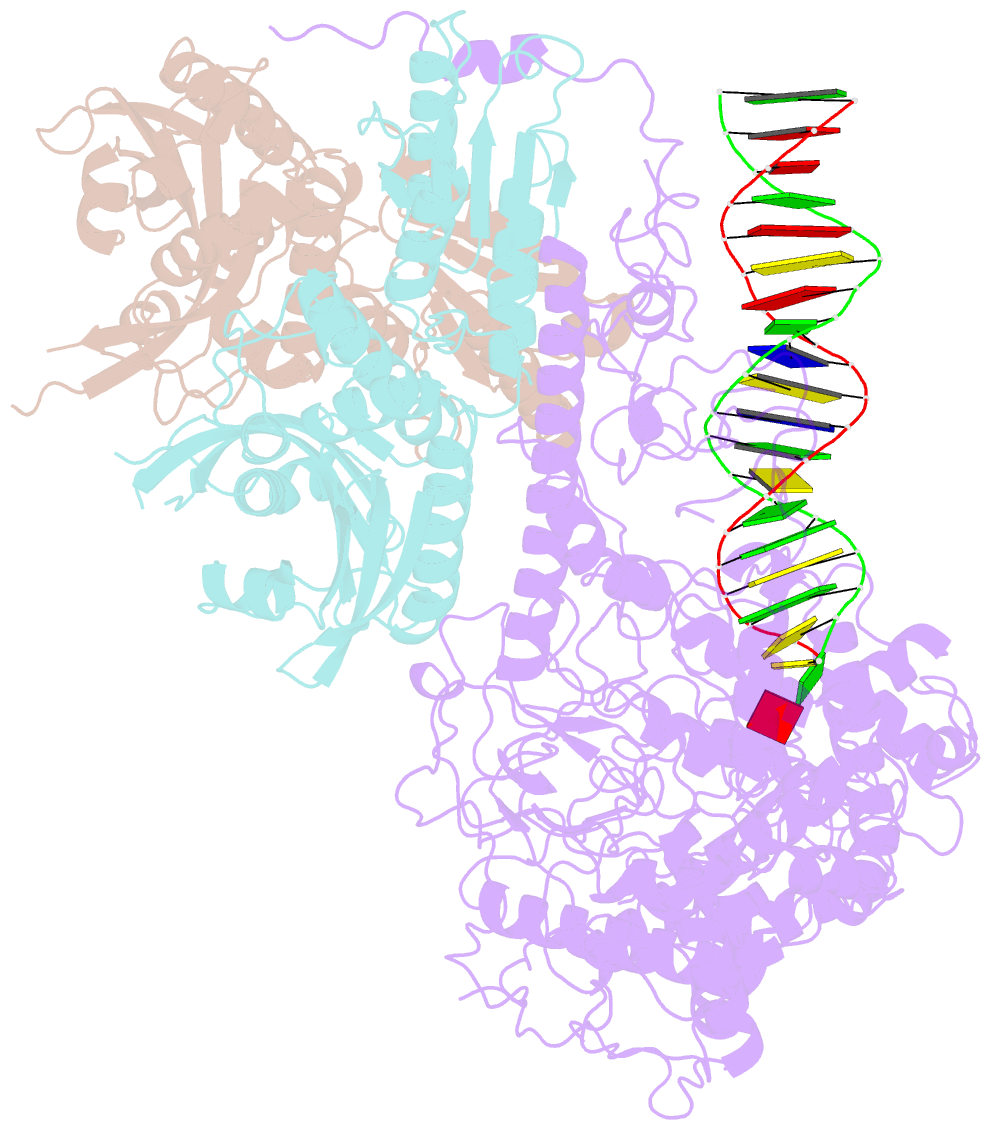
- cryo-EM structure of the primer separation complex (ix) of human mitochondrial DNA polymerase gamma
- Reference
- Buchel G, Nayak AR, Herbine K, Sarfallah A, Sokolova VO, Zamudio-Ochoa A, Temiakov D (2023): "Structural basis for DNA proofreading." Nat Commun, 14, 8501. doi: 10.1038/s41467-023-44198-8.
- Abstract
- DNA polymerase (DNAP) can correct errors in DNA during replication by proofreading, a process critical for cell viability. However, the mechanism by which an erroneously incorporated base translocates from the polymerase to the exonuclease site and the corrected DNA terminus returns has remained elusive. Here, we present an ensemble of nine high-resolution structures representing human mitochondrial DNA polymerase Gamma, Polγ, captured during consecutive proofreading steps. The structures reveal key events, including mismatched base recognition, its dissociation from the polymerase site, forward translocation of DNAP, alterations in DNA trajectory, repositioning and refolding of elements for primer separation, DNAP backtracking, and displacement of the mismatched base into the exonuclease site. Altogether, our findings suggest a conserved 'bolt-action' mechanism of proofreading based on iterative cycles of DNAP translocation without dissociation from the DNA, facilitating primer transfer between catalytic sites. Functional assays and mutagenesis corroborate this mechanism, connecting pathogenic mutations to crucial structural elements in proofreading steps.