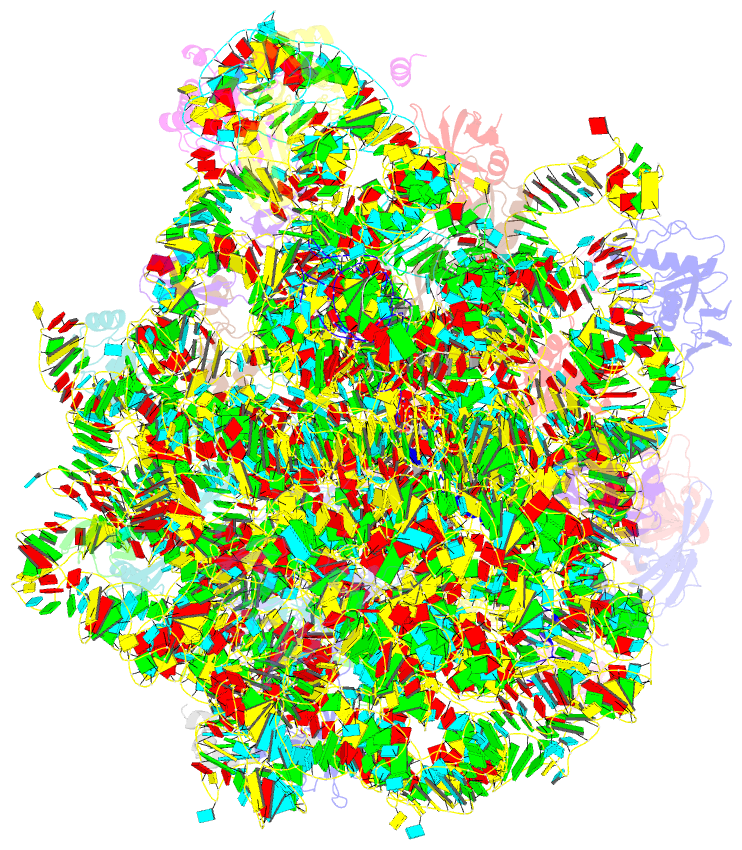
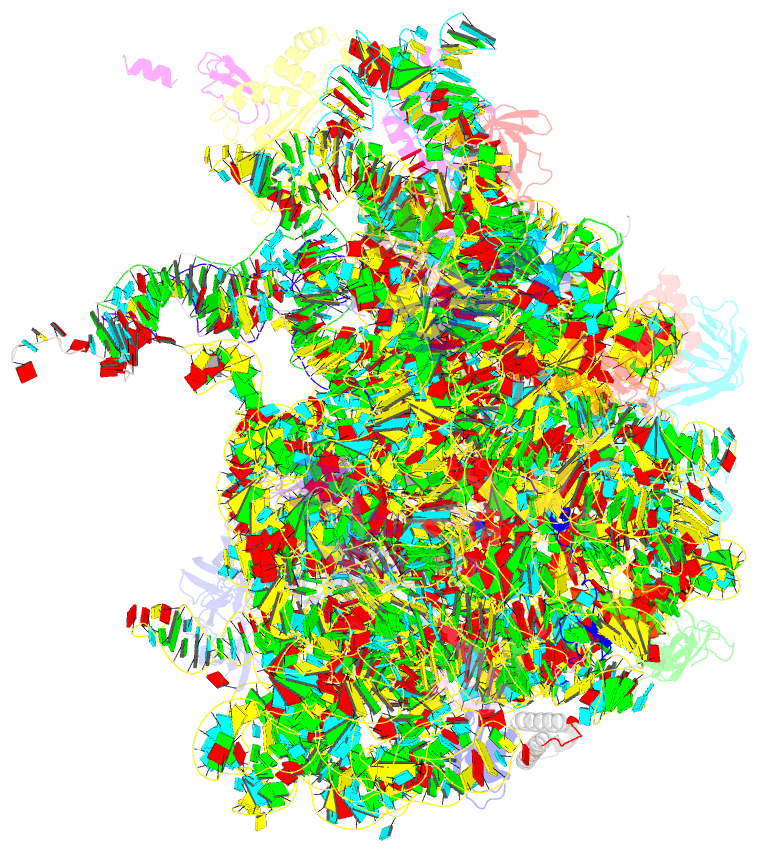
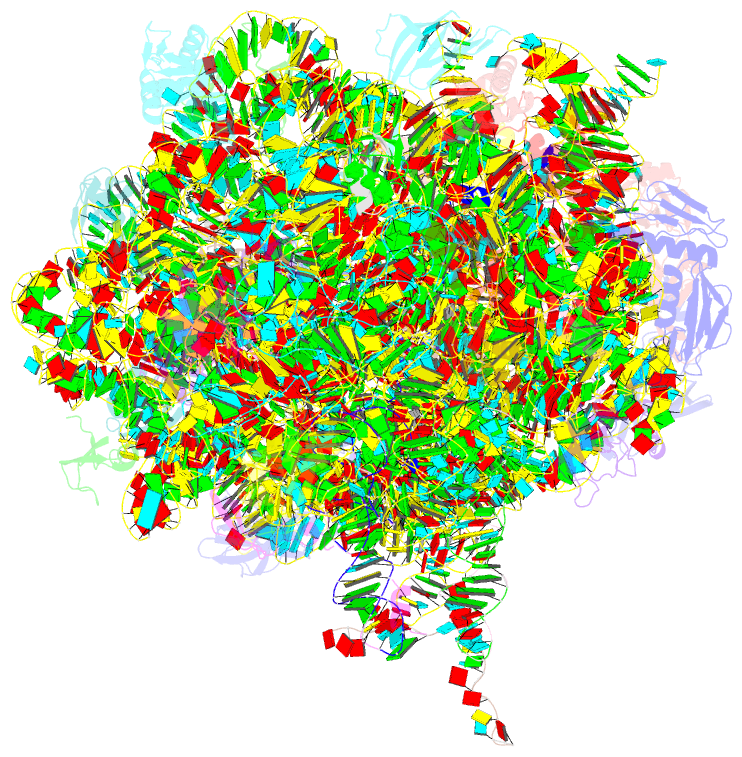
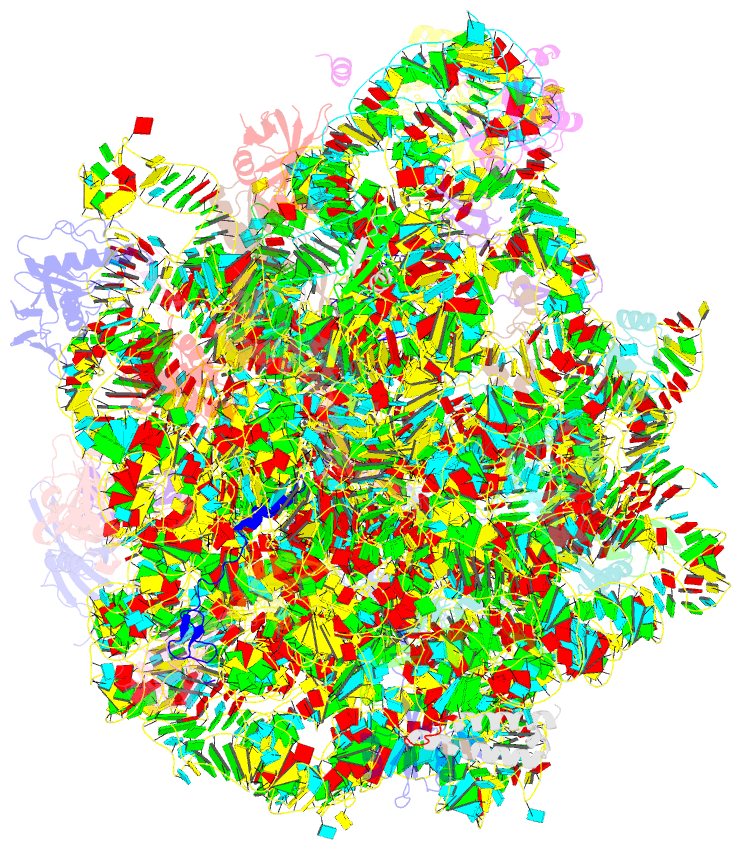
Summary information and primary citation
- PDB-id
- 8g6y; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ribosome
- Method
- cryo-EM (2.09 Å)
- Summary
- Structure of wt e.coli ribosome 50s subunit with complexed with mrna, p-site fmet-nh-trnafmet and a-site 3-aminopyridine-4-carboxylic acid charged nh-trnaphe
- Reference
- Majumdar C, Walker JA, Francis MB, Schepartz A, Cate JHD (2023): "Aminobenzoic Acid Derivatives Obstruct Induced Fit in the Catalytic Center of the Ribosome." Acs Cent.Sci., 9, 1160-1169. doi: 10.1021/acscentsci.3c00153.
- Abstract
- The Escherichia coli (E. coli) ribosome can incorporate a variety of non-l-α-amino acid monomers into polypeptide chains in vitro but with poor efficiency. Although these monomers span a diverse set of compounds, there exists no high-resolution structural information regarding their positioning within the catalytic center of the ribosome, the peptidyl transferase center (PTC). Thus, details regarding the mechanism of amide bond formation and the structural basis for differences and defects in incorporation efficiency remain unknown. Within a set of three aminobenzoic acid derivatives-3-aminopyridine-4-carboxylic acid (Apy), ortho-aminobenzoic acid (oABZ), and meta-aminobenzoic acid (mABZ)-the ribosome incorporates Apy into polypeptide chains with the highest efficiency, followed by oABZ and then mABZ, a trend that does not track with the nucleophilicity of the reactive amines. Here, we report high-resolution cryo-EM structures of the ribosome with each of these three aminobenzoic acid derivatives charged on tRNA bound in the aminoacyl-tRNA site (A-site). The structures reveal how the aromatic ring of each monomer sterically blocks the positioning of nucleotide U2506, thereby preventing rearrangement of nucleotide U2585 and the resulting induced fit in the PTC required for efficient amide bond formation. They also reveal disruptions to the bound water network that is believed to facilitate formation and breakdown of the tetrahedral intermediate. Together, the cryo-EM structures reported here provide a mechanistic rationale for differences in reactivity of aminobenzoic acid derivatives relative to l-α-amino acids and each other and identify stereochemical constraints on the size and geometry of non-monomers that can be accepted efficiently by wild-type ribosomes.