Summary information and primary citation
- PDB-id
- 8h1t; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- gene regulation
- Method
- cryo-EM (3.0 Å)
- Summary
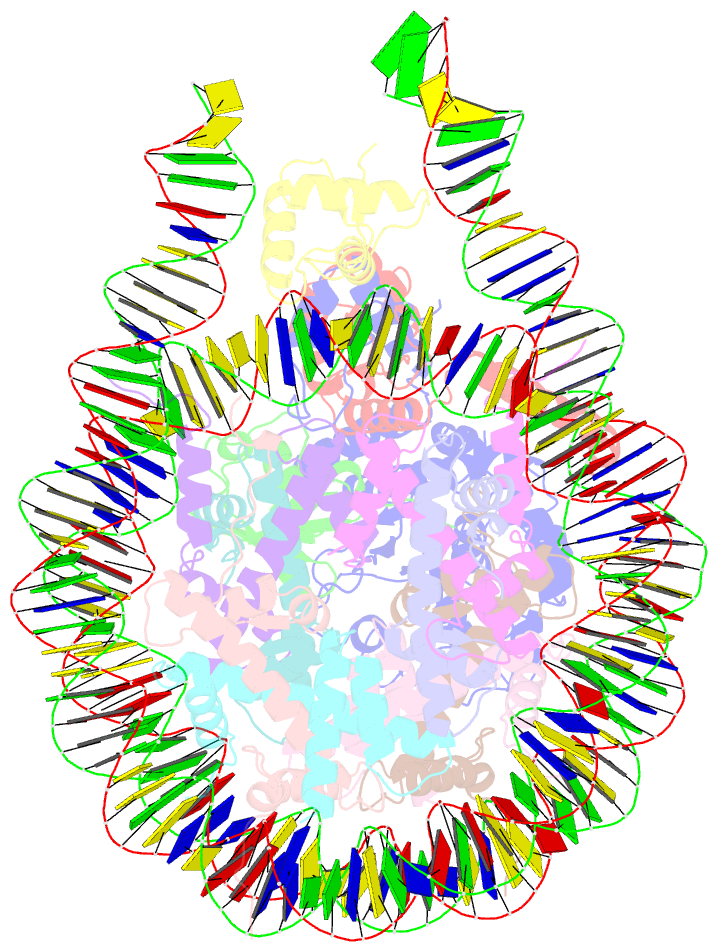
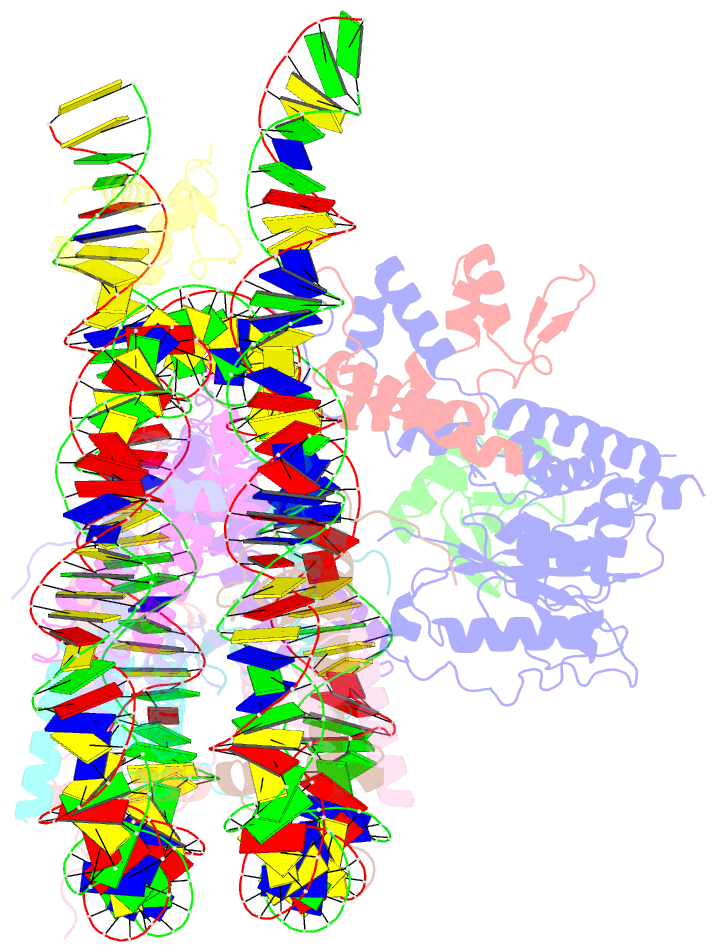
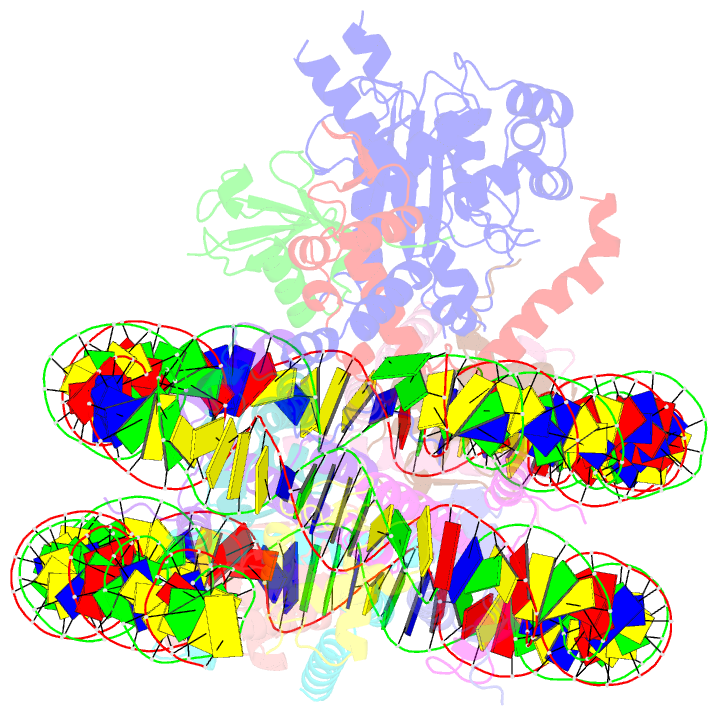
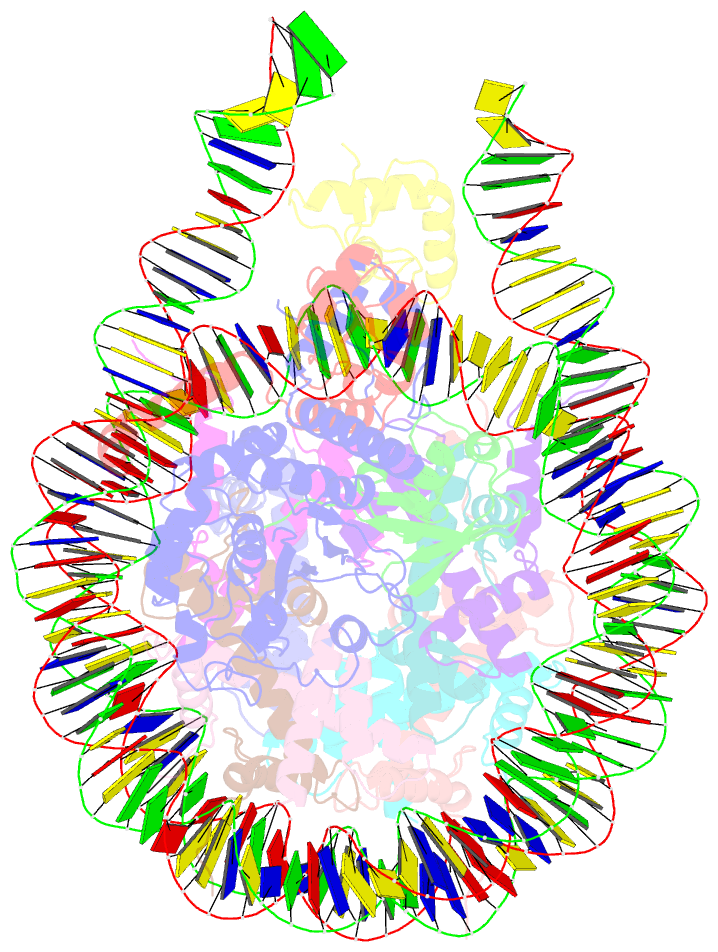
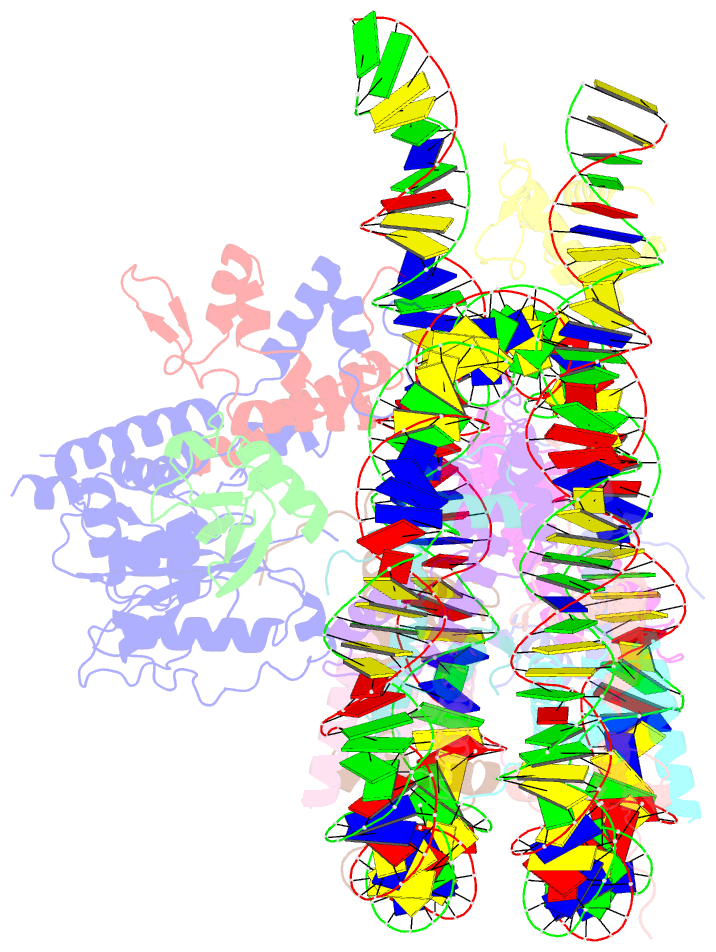
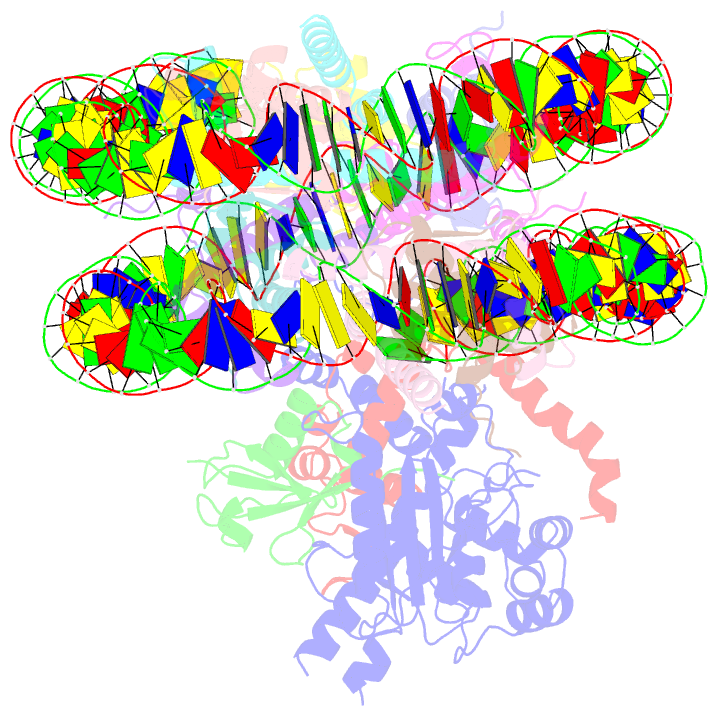
- cryo-EM structure of bap1-asxl1 bound to chromatosome
- Reference
- Ge W, Yu C, Li J, Yu Z, Li X, Zhang Y, Liu CP, Li Y, Tian C, Zhang X, Li G, Zhu B, Xu RM (2023): "Basis of the H2AK119 specificity of the Polycomb repressive deubiquitinase." Nature, 616, 176-182. doi: 10.1038/s41586-023-05841-y.
- Abstract
- Repression of gene expression by protein complexes of the Polycomb group is a fundamental mechanism that governs embryonic development and cell-type specification1-3. The Polycomb repressive deubiquitinase (PR-DUB) complex removes the ubiquitin moiety from monoubiquitinated histone H2A K119 (H2AK119ub1) on the nucleosome4, counteracting the ubiquitin E3 ligase activity of Polycomb repressive complex 1 (PRC1)5 to facilitate the correct silencing of genes by Polycomb proteins and safeguard active genes from inadvertent silencing by PRC1 (refs. 6-9). The intricate biological function of PR-DUB requires accurate targeting of H2AK119ub1, but PR-DUB can deubiquitinate monoubiquitinated free histones and peptide substrates indiscriminately; the basis for its exquisite nucleosome-dependent substrate specificity therefore remains unclear. Here we report the cryo-electron microscopy structure of human PR-DUB, composed of BAP1 and ASXL1, in complex with the chromatosome. We find that ASXL1 directs the binding of the positively charged C-terminal extension of BAP1 to nucleosomal DNA and histones H3-H4 near the dyad, an addition to its role in forming the ubiquitin-binding cleft. Furthermore, a conserved loop segment of the catalytic domain of BAP1 is situated near the H2A-H2B acidic patch. This distinct nucleosome-binding mode displaces the C-terminal tail of H2A from the nucleosome surface, and endows PR-DUB with the specificity for H2AK119ub1.