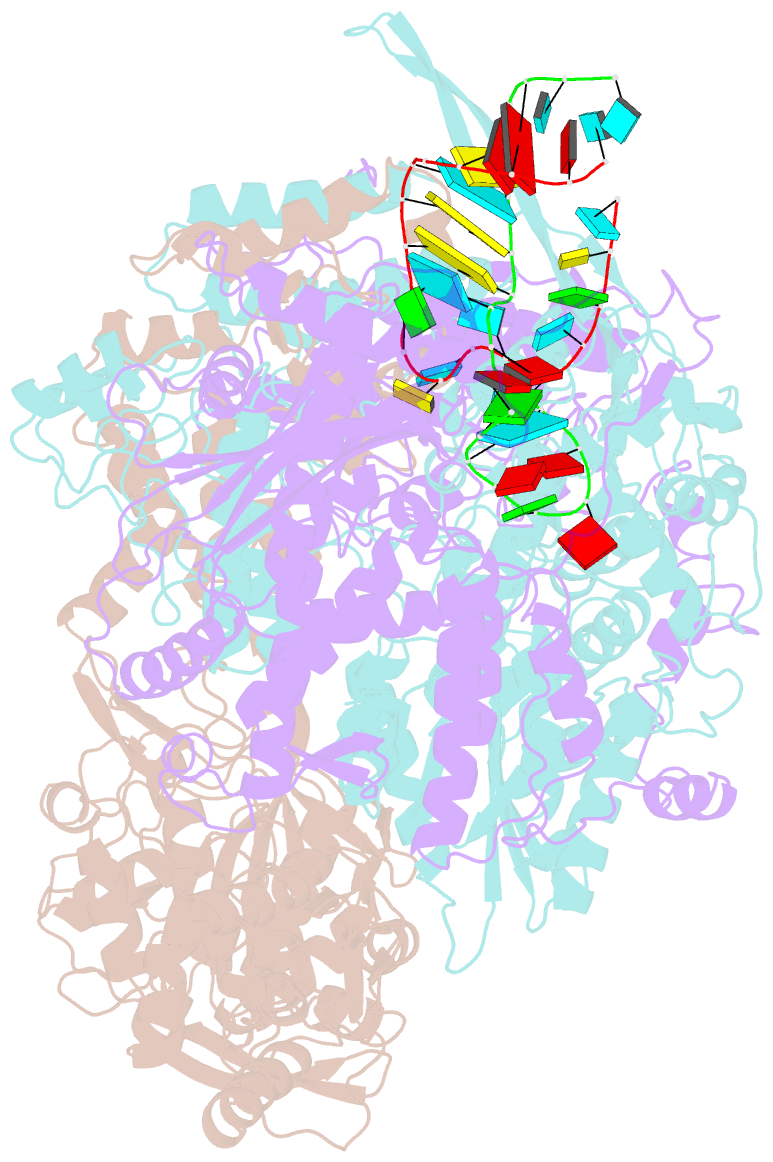
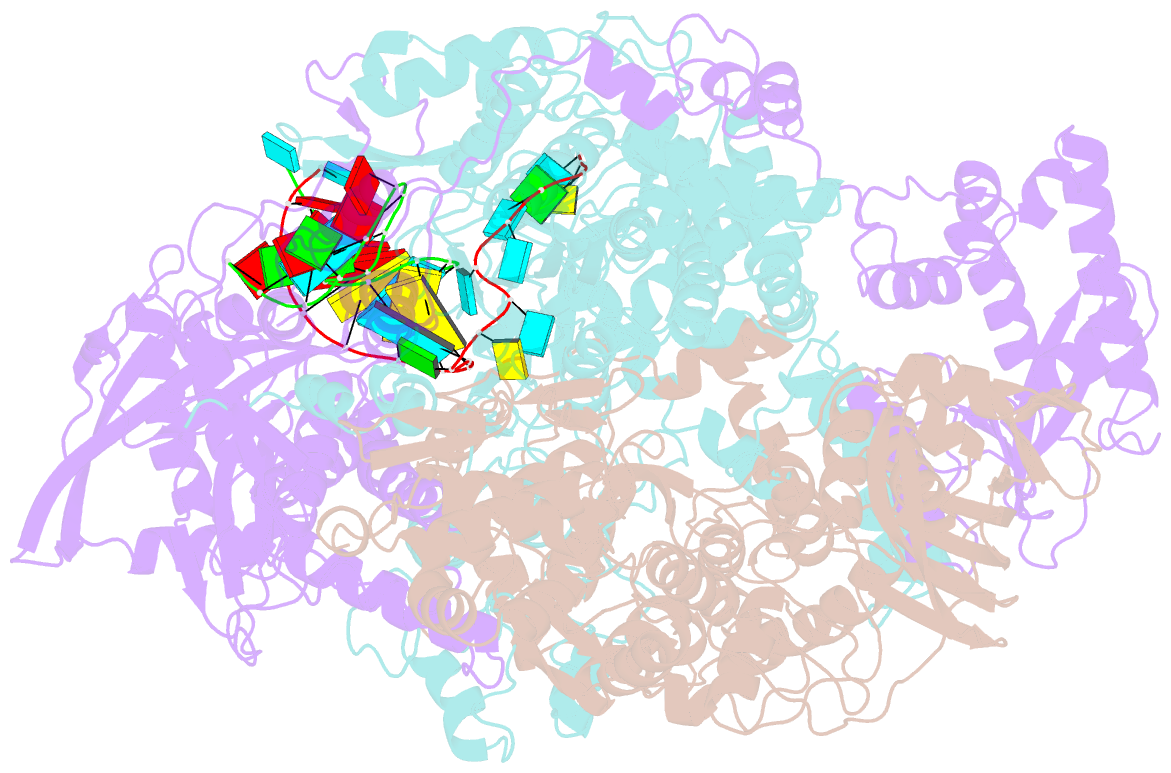
Summary information and primary citation
- PDB-id
- 8h69; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein
- Method
- cryo-EM (3.7 Å)
- Summary
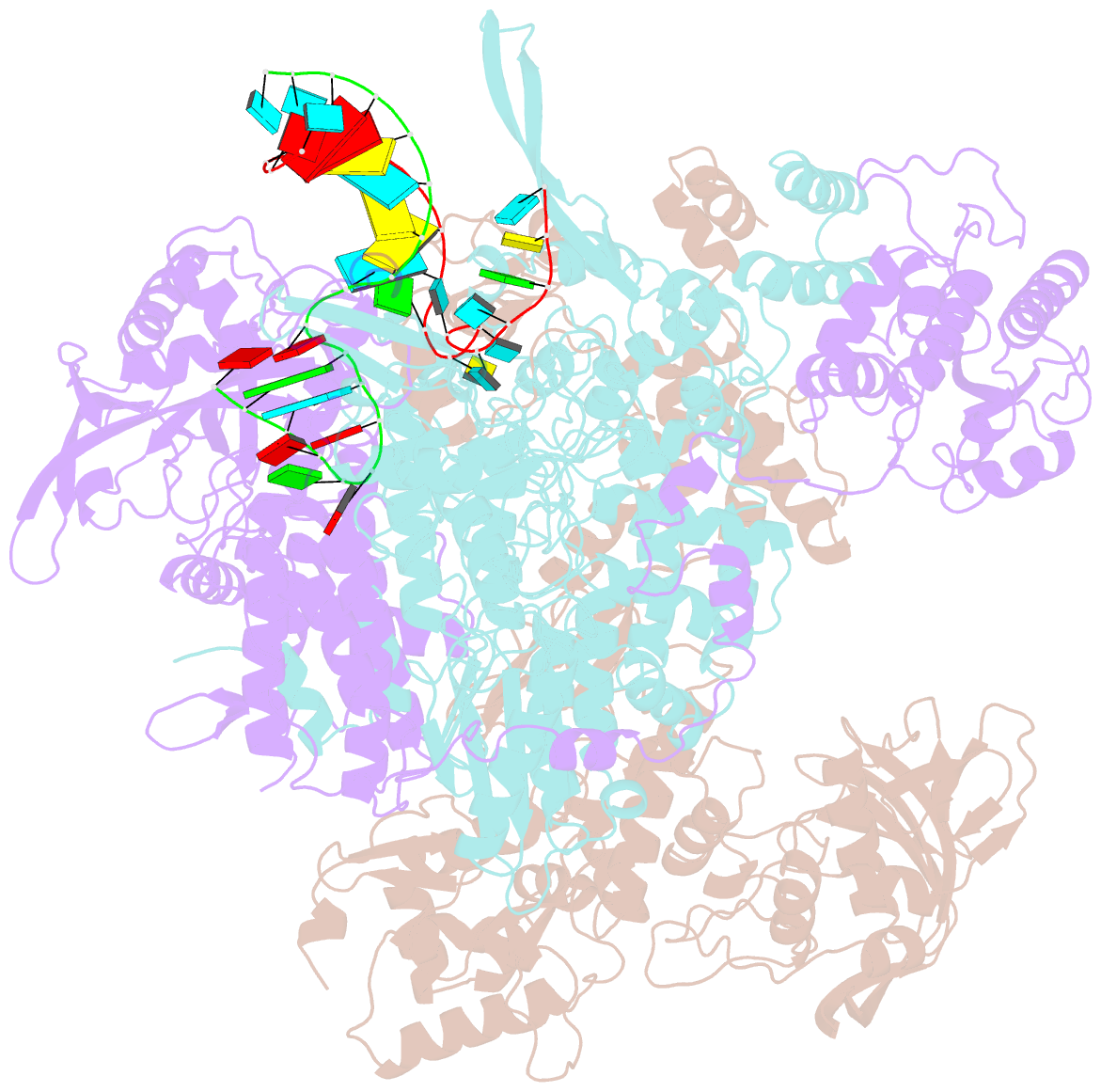
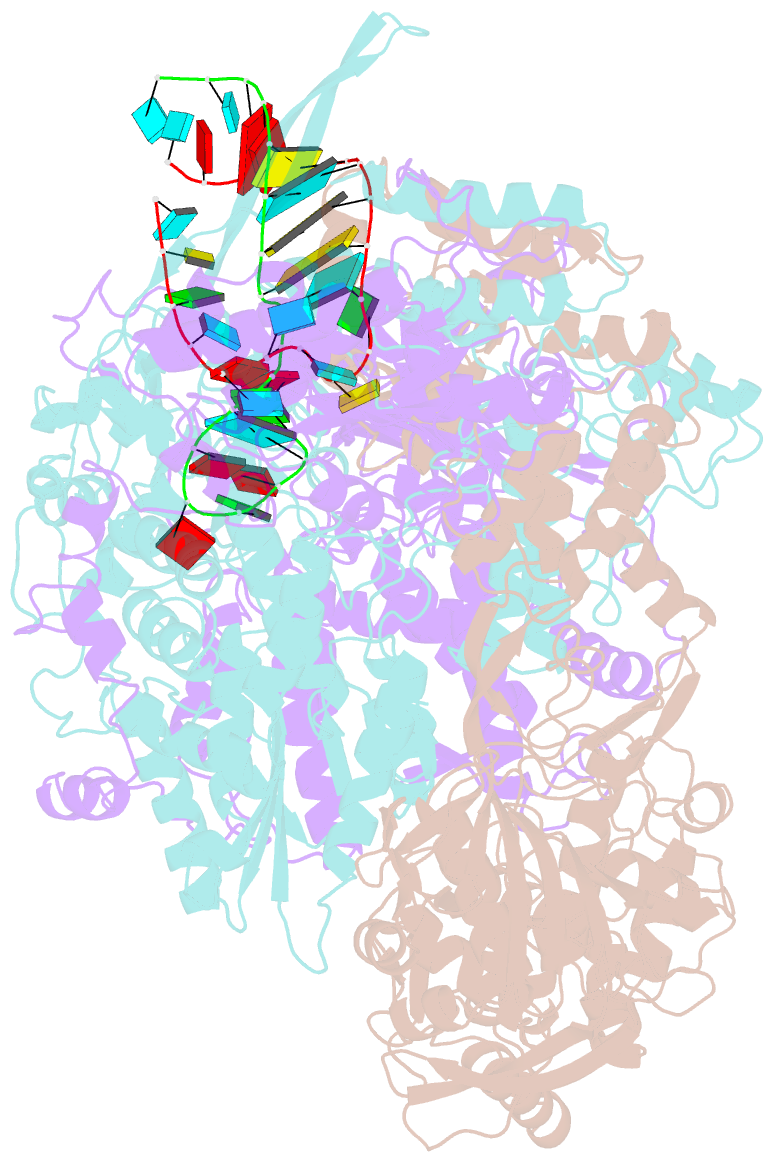
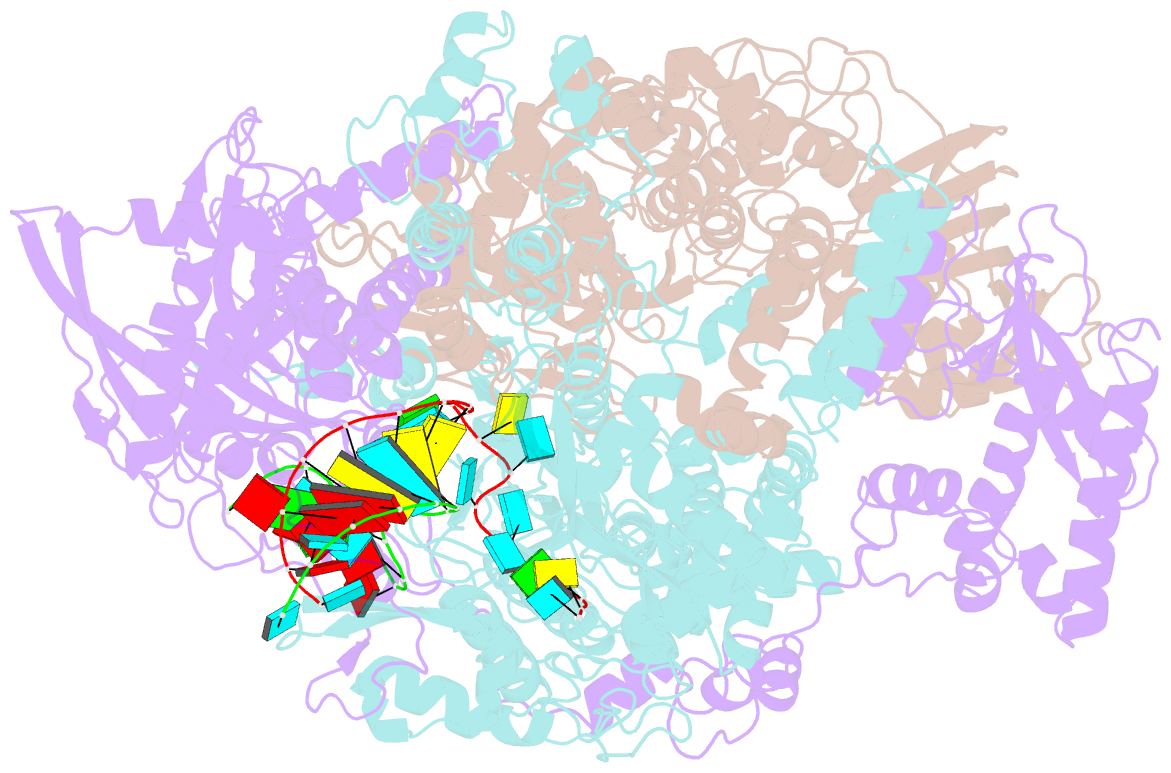
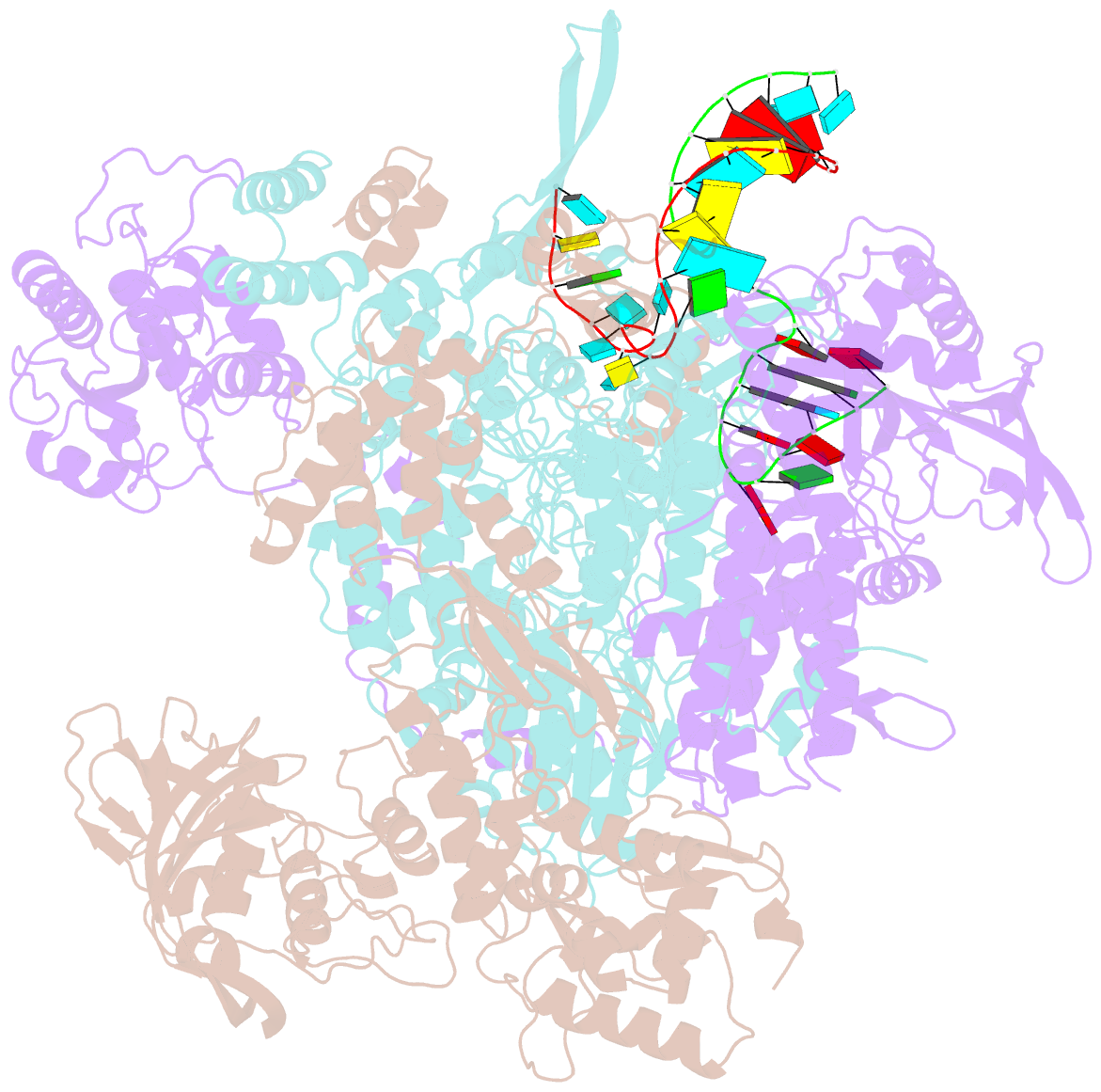
- cryo-EM structure of influenza RNA polymerase
- Reference
- Li H, Wu Y, Li M, Guo L, Gao Y, Wang Q, Zhang J, Lai Z, Zhang X, Zhu L, Lan P, Rao Z, Liu Y, Liang H (2023): "An intermediate state allows influenza polymerase to switch smoothly between transcription and replication cycles." Nat.Struct.Mol.Biol., 30, 1183-1192. doi: 10.1038/s41594-023-01043-2.
- Abstract
- Influenza polymerase (FluPol) transcribes viral mRNA at the beginning of the viral life cycle and initiates genome replication after viral protein synthesis. However, it remains poorly understood how FluPol switches between its transcription and replication states, especially given that the structural bases of these two functions are fundamentally different. Here we propose a mechanism by which FluPol achieves functional switching between these two states through a previously unstudied conformation, termed an 'intermediate state'. Using cryo-electron microscopy, we obtained a structure of the intermediate state of H5N1 FluPol at 3.7 Å, which is characterized by a blocked cap-binding domain and a contracted core region. Structural analysis results suggest that the intermediate state may allow FluPol to transition smoothly into either the transcription or replication state. Furthermore, we show that the formation of the intermediate state is required for both the transcription and replication activities of FluPol, leading us to conclude that the transcription and replication cycles of FluPol are regulated via this intermediate state.