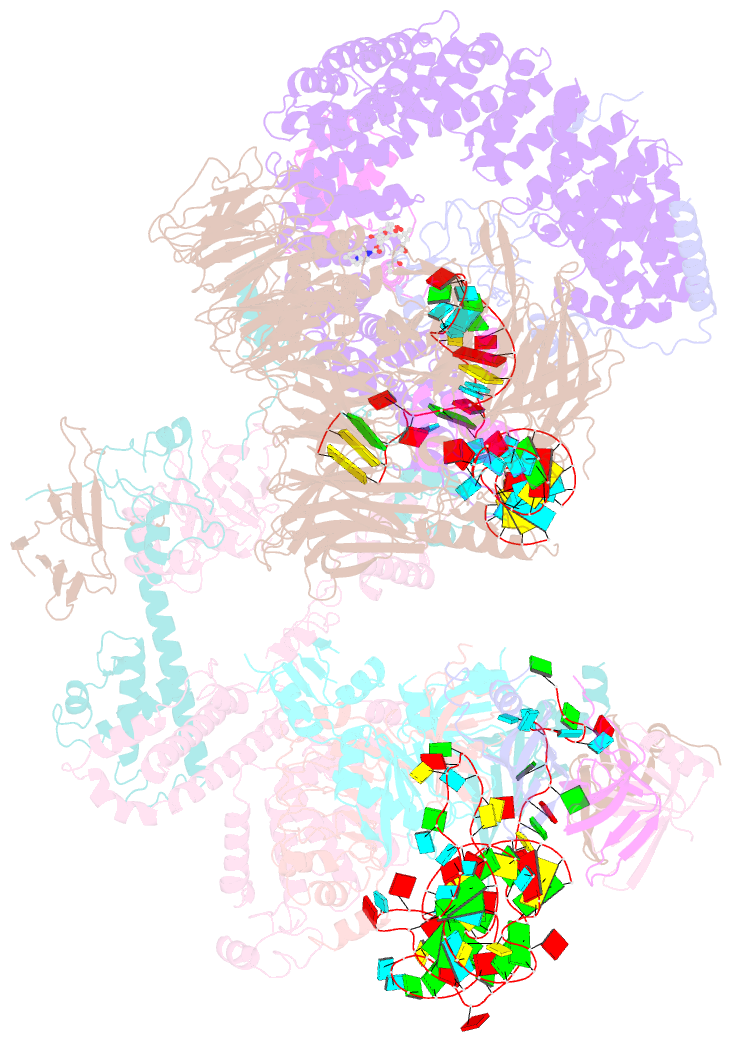
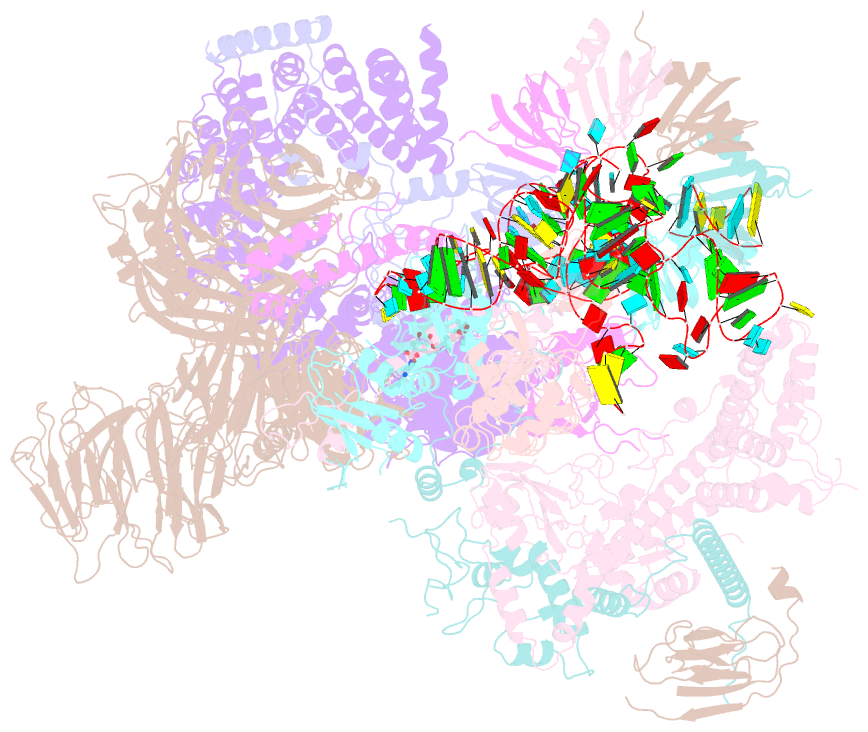
Summary information and primary citation
- PDB-id
- 8hk1; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- splicing
- Method
- cryo-EM (2.7 Å)
- Summary
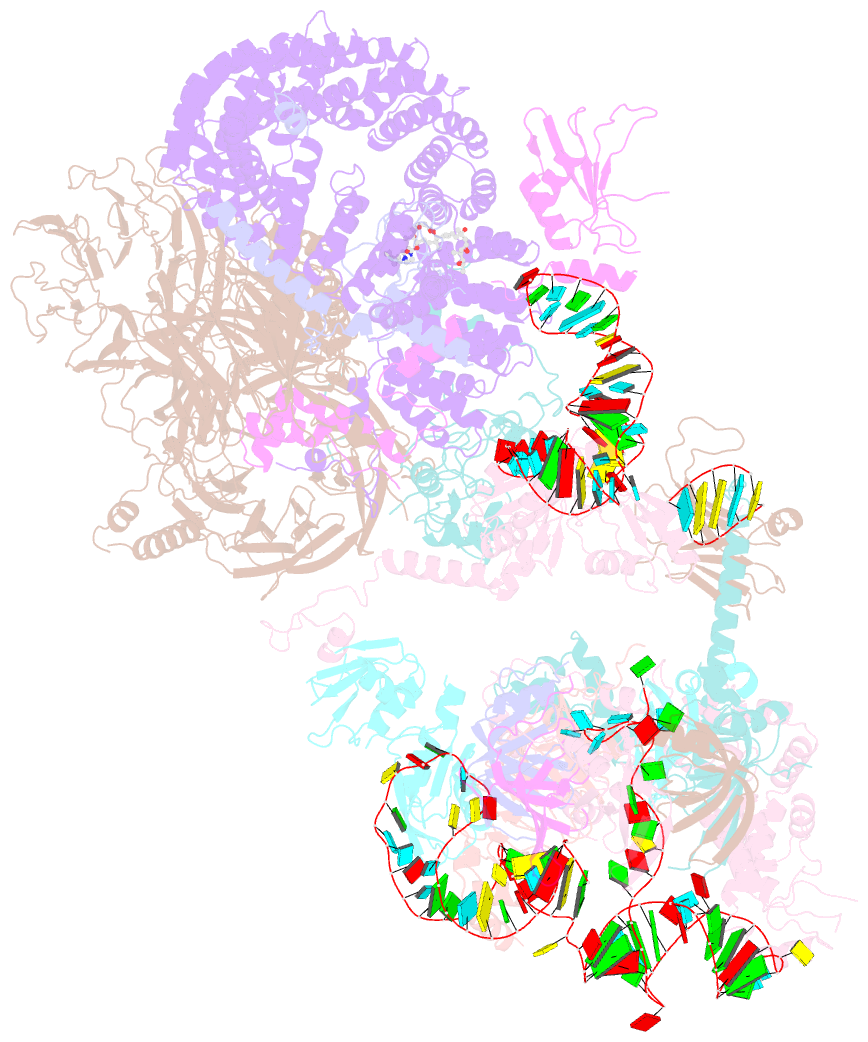
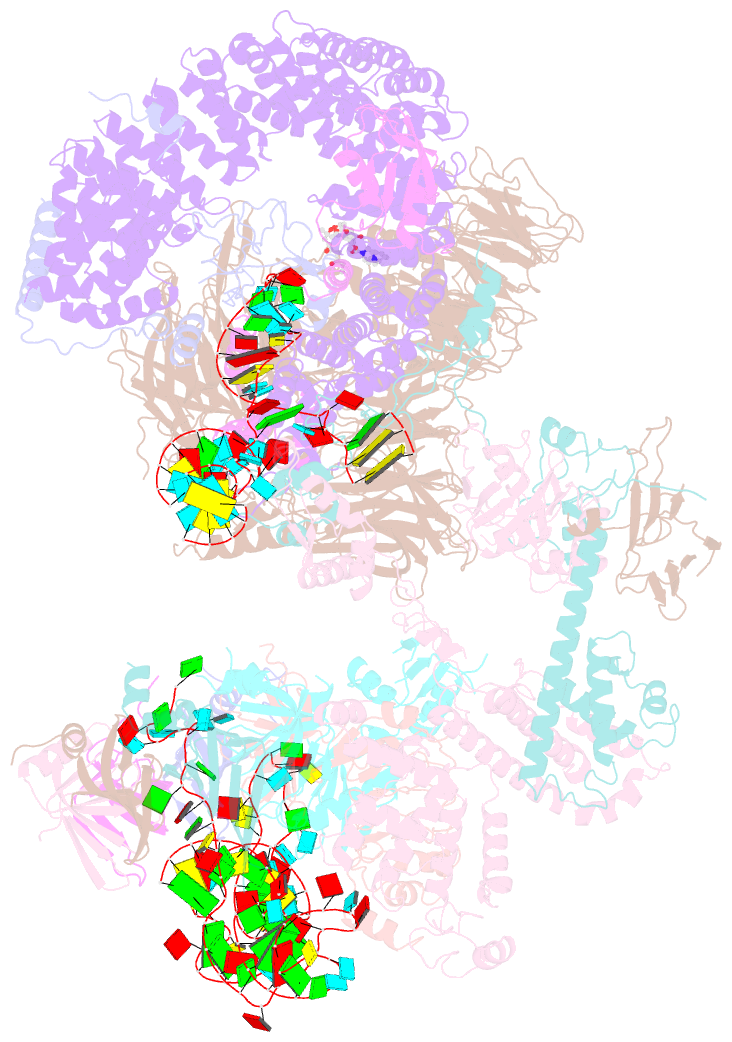
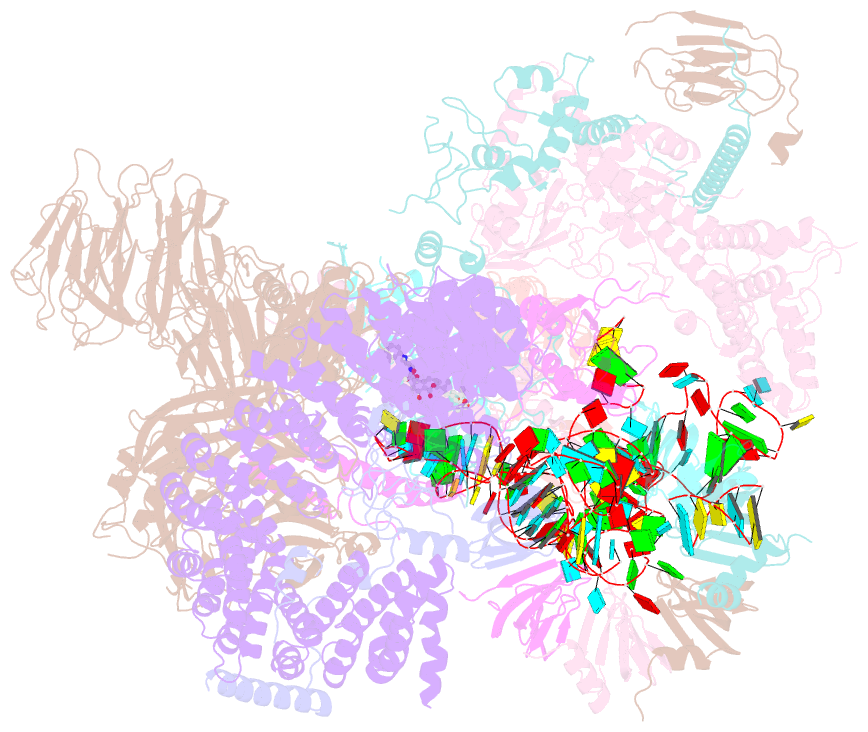
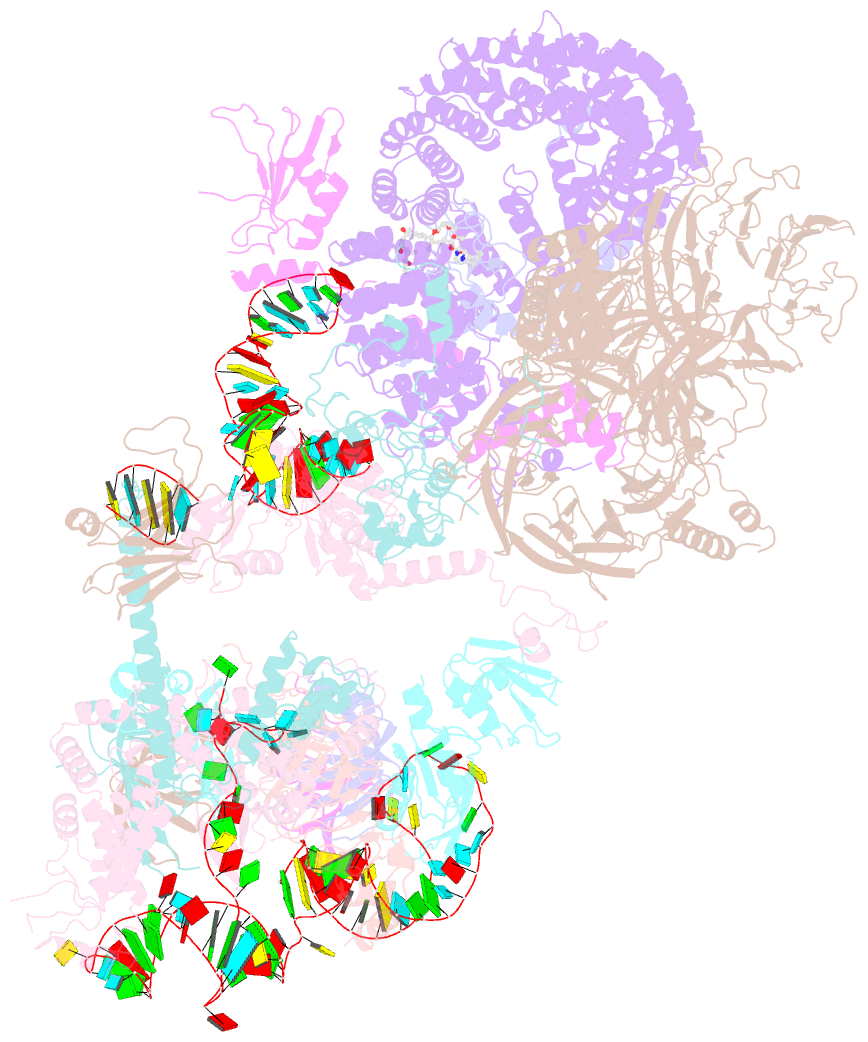
- The cryo-EM structure of human pre-17s u2 snrnp
- Reference
- Yang F, Bian T, Zhan X, Chen Z, Xing Z, Larsen NA, Zhang X, Shi Y (2023): "Mechanisms of the RNA helicases DDX42 and DDX46 in human U2 snRNP assembly." Nat Commun, 14, 897. doi: 10.1038/s41467-023-36489-x.
- Abstract
- Three RNA helicases - DDX42, DDX46 and DHX15 - are found to be associated with human U2 snRNP, but their roles and mechanisms in U2 snRNP and spliceosome assembly are insufficiently understood. Here we report the cryo-electron microscopy (cryo-EM) structures of the DDX42-SF3b complex and a putative assembly precursor of 17S U2 snRNP that contains DDX42 (DDX42-U2 complex). DDX42 is anchored on SF3B1 through N-terminal sequences, with its N-plug occupying the RNA path of SF3B1. The binding mode of DDX42 to SF3B1 is in striking analogy to that of DDX46. In the DDX42-U2 complex, the N-terminus of DDX42 remains anchored on SF3B1, but the helicase domain has been displaced by U2 snRNA and TAT-SF1. Through in vitro assays, we show DDX42 and DDX46 are mutually exclusive in terms of binding to SF3b. Cancer-driving mutations of SF3B1 target the residues in the RNA path that directly interact with DDX42 and DDX46. These findings reveal the distinct roles of DDX42 and DDX46 in assembly of 17S U2 snRNP and provide insights into the mechanisms of SF3B1 cancer mutations.