Summary information and primary citation
- PDB-id
- 8hud; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA-DNA
- Method
- cryo-EM (3.43 Å)
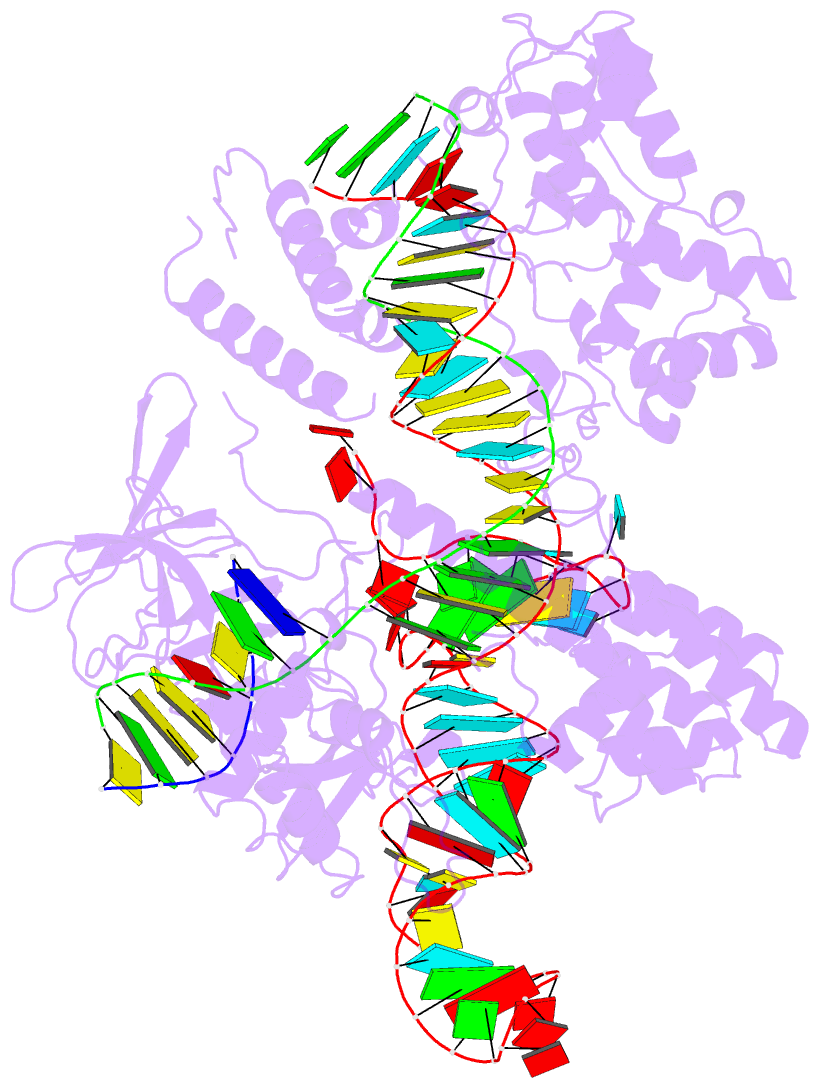
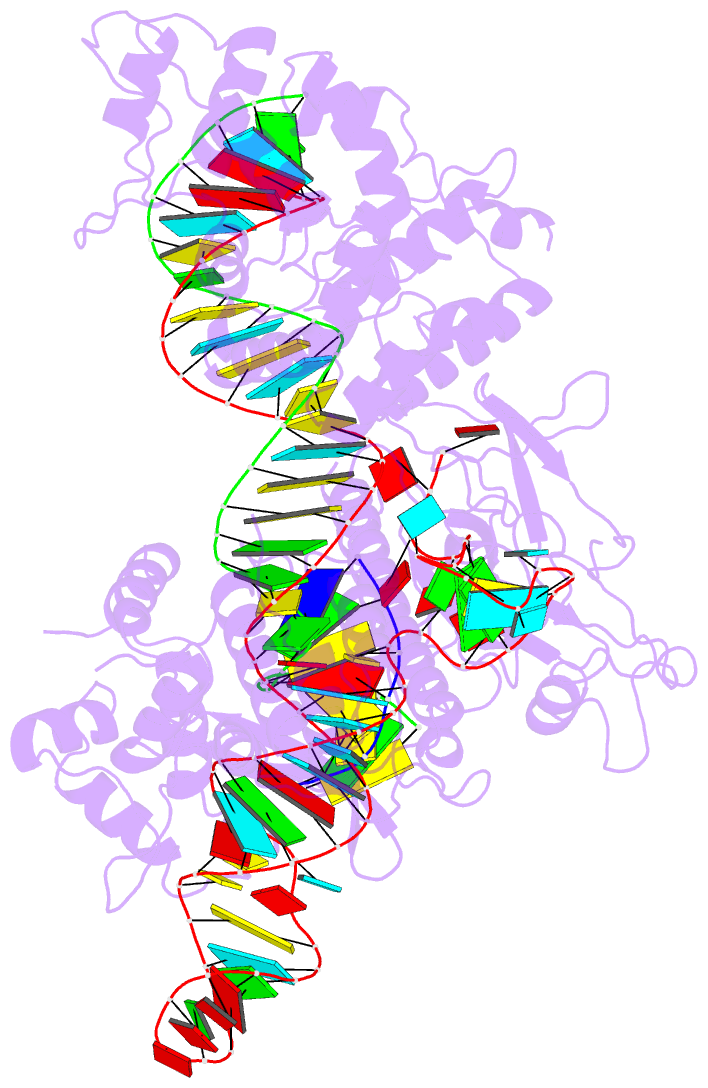
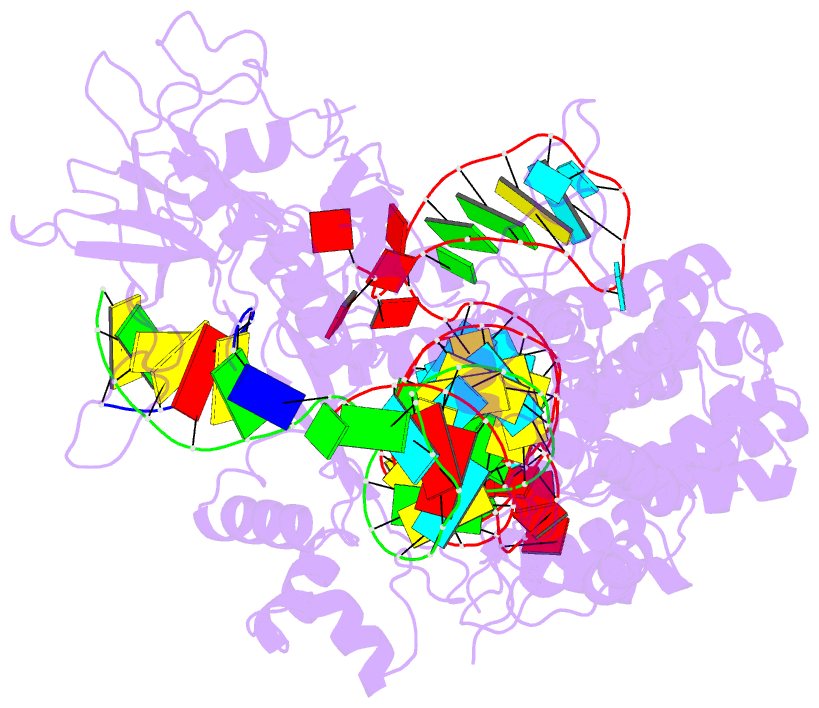
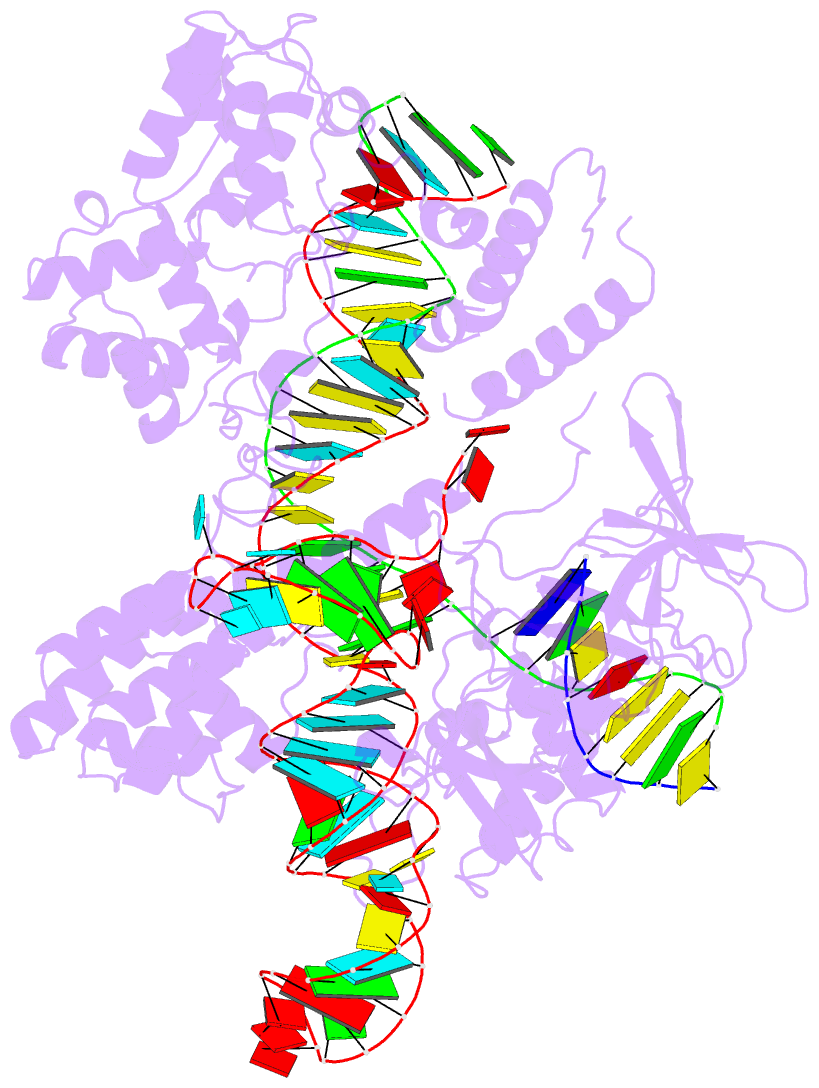
- Summary
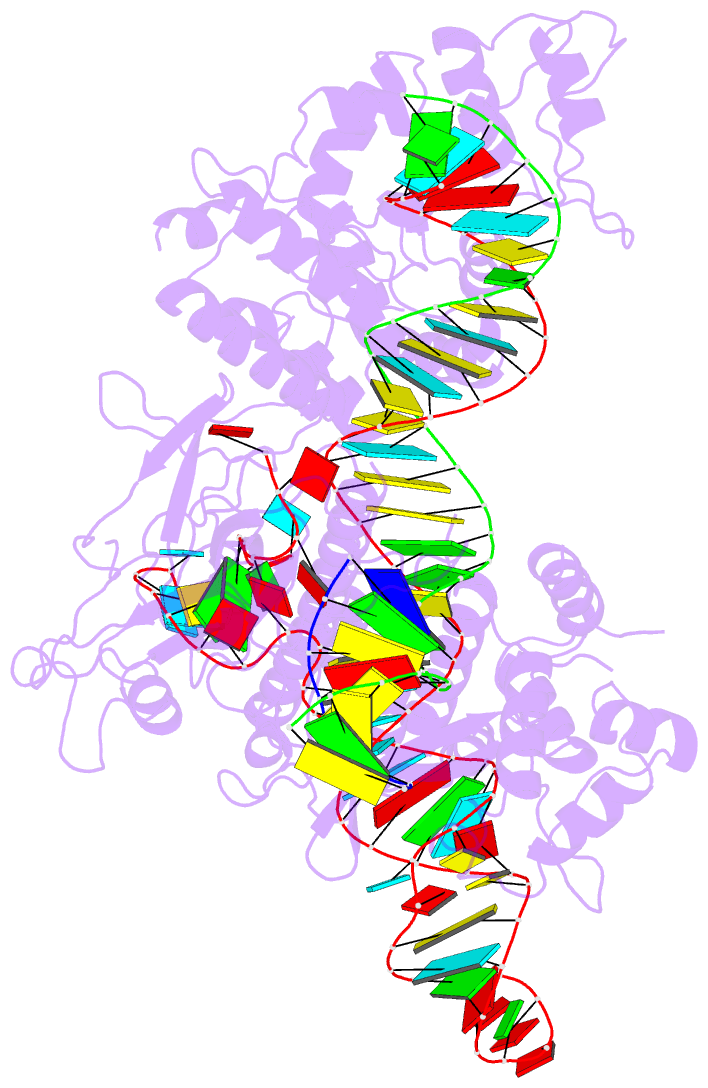
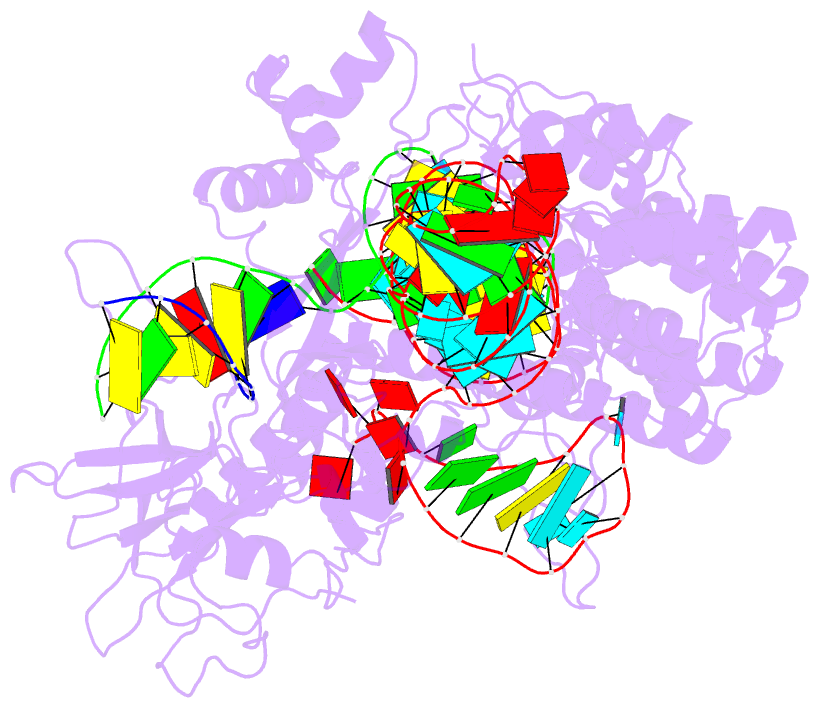
- cryo-EM structure of the evcas9-sgrna-target DNA ternary complex
- Reference
- Tang N, Wu Z, Gao Y, Chen W, Wang Z, Su M, Ji W, Ji Q (2024): "Molecular Basis and Genome Editing Applications of a Compact Eubacterium ventriosum CRISPR-Cas9 System." Acs Synth Biol, 13, 269-281. doi: 10.1021/acssynbio.3c00501.
- Abstract
- CRISPR-Cas9 systems have been widely harnessed for diverse genome editing applications because of their ease of use and high efficiency. However, the large molecular sizes and strict PAM requirements of commonly used CRISPR-Cas9 systems restrict their broad applications in therapeutics. Here, we report the molecular basis and genome editing applications of a novel compact type II-A Eubacterium ventriosum CRISPR-Cas9 system (EvCas9) with 1107 residues and distinct 5'-NNGDGN-3' (where D represents A, T, or G) PAM specificity. We determine the cryo-EM structure of EvCas9 in a complex with an sgRNA and a target DNA, revealing the detailed PAM recognition and sgRNA and target DNA association mechanisms. Additionally, we demonstrate the robust genome editing capacity of EvCas9 in bacteria and human cells with superior fidelity compared to SaCas9 and SpCas9, and we engineer it to be efficient base editors by fusing a cytidine or adenosine deaminase. Collectively, our results facilitate further understanding of CRISPR-Cas9 working mechanisms and expand the compact CRISPR-Cas9 toolbox.