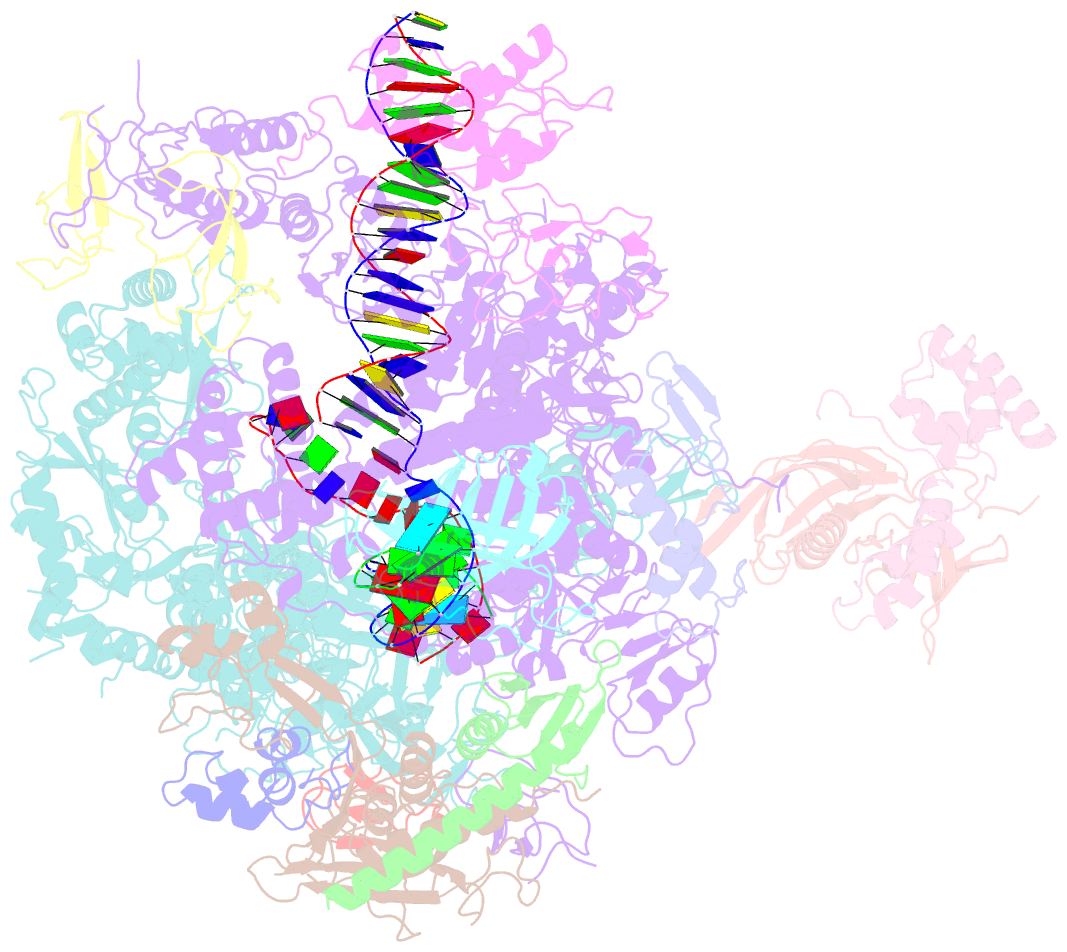
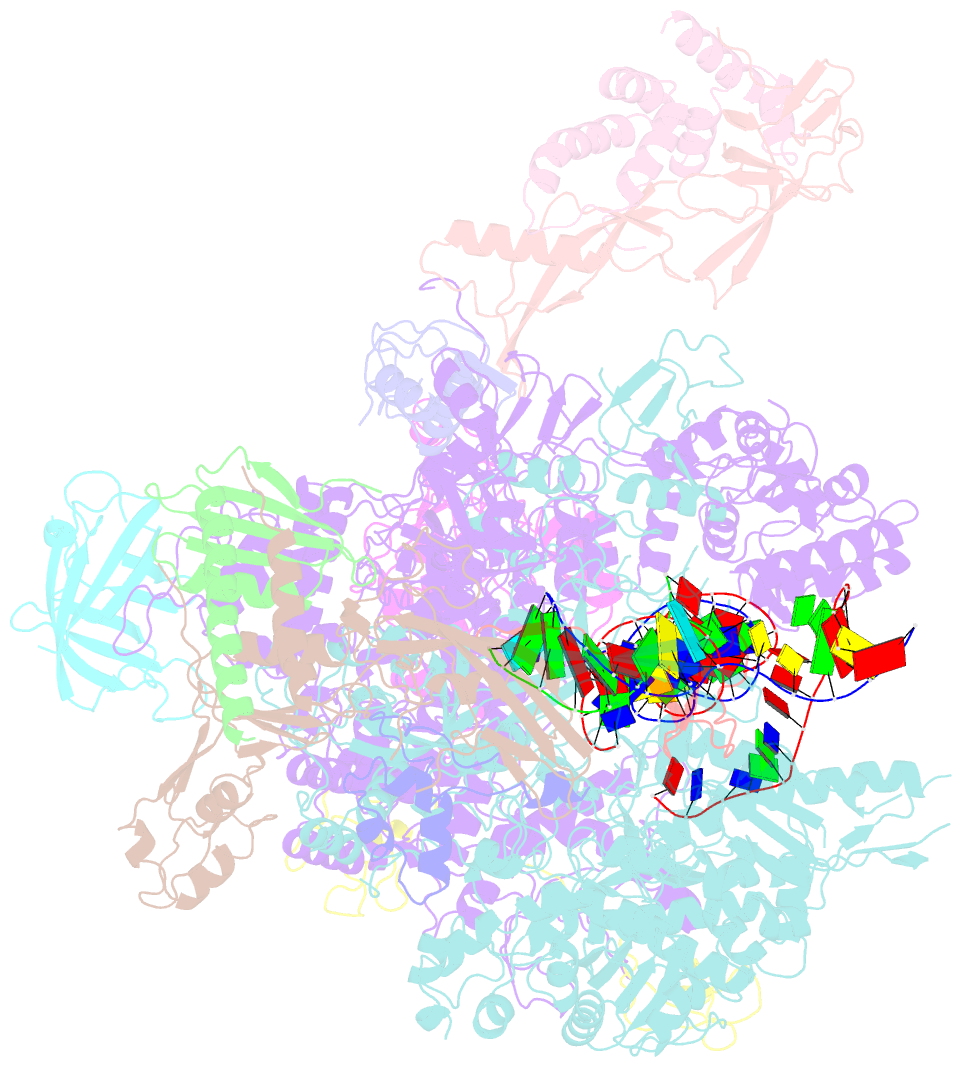
Summary information and primary citation
- PDB-id
- 8hyj; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- cryo-EM (4.3 Å)
- Summary
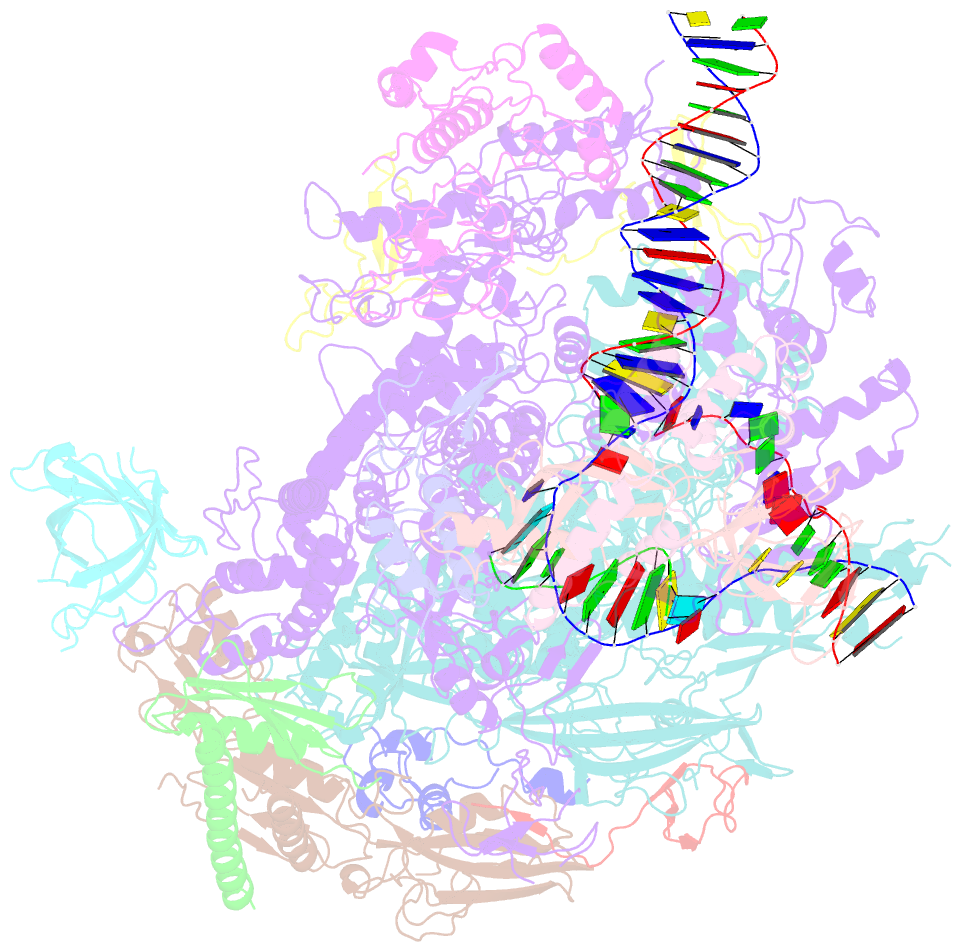
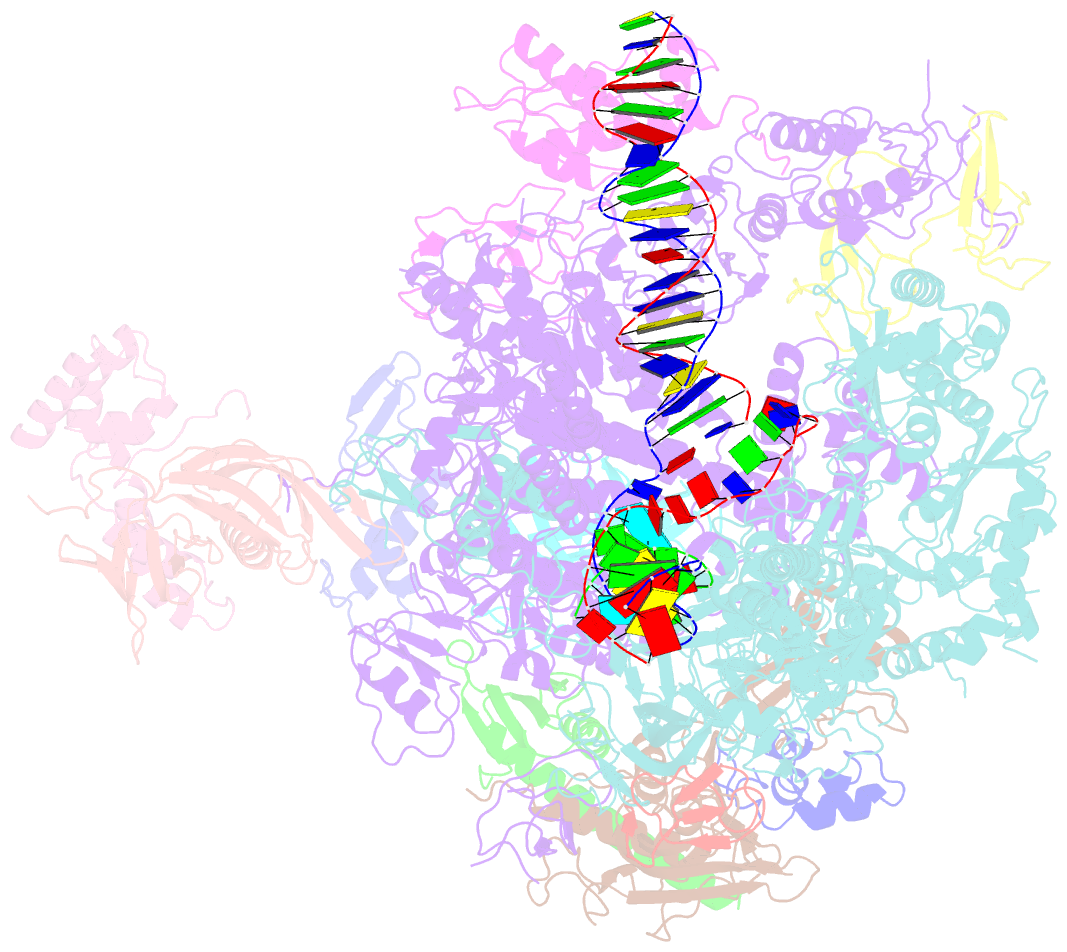
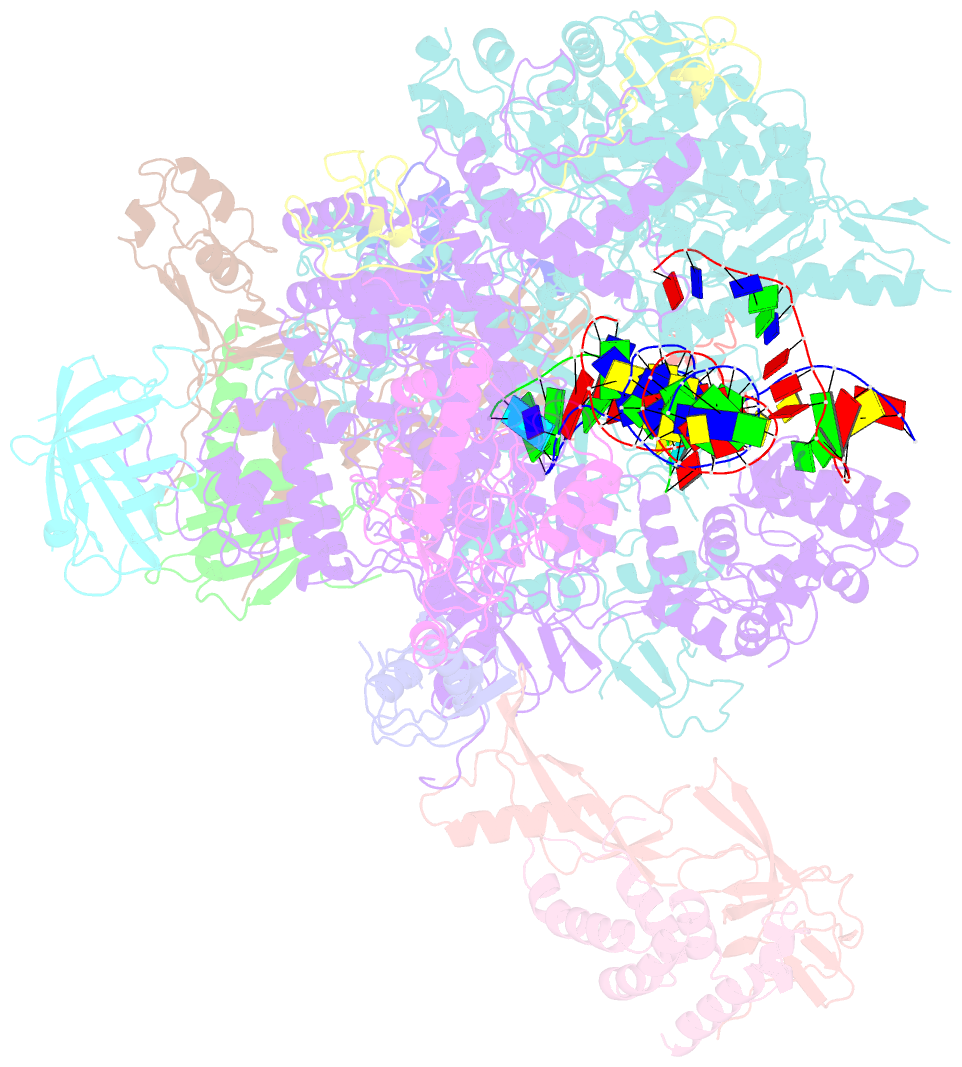
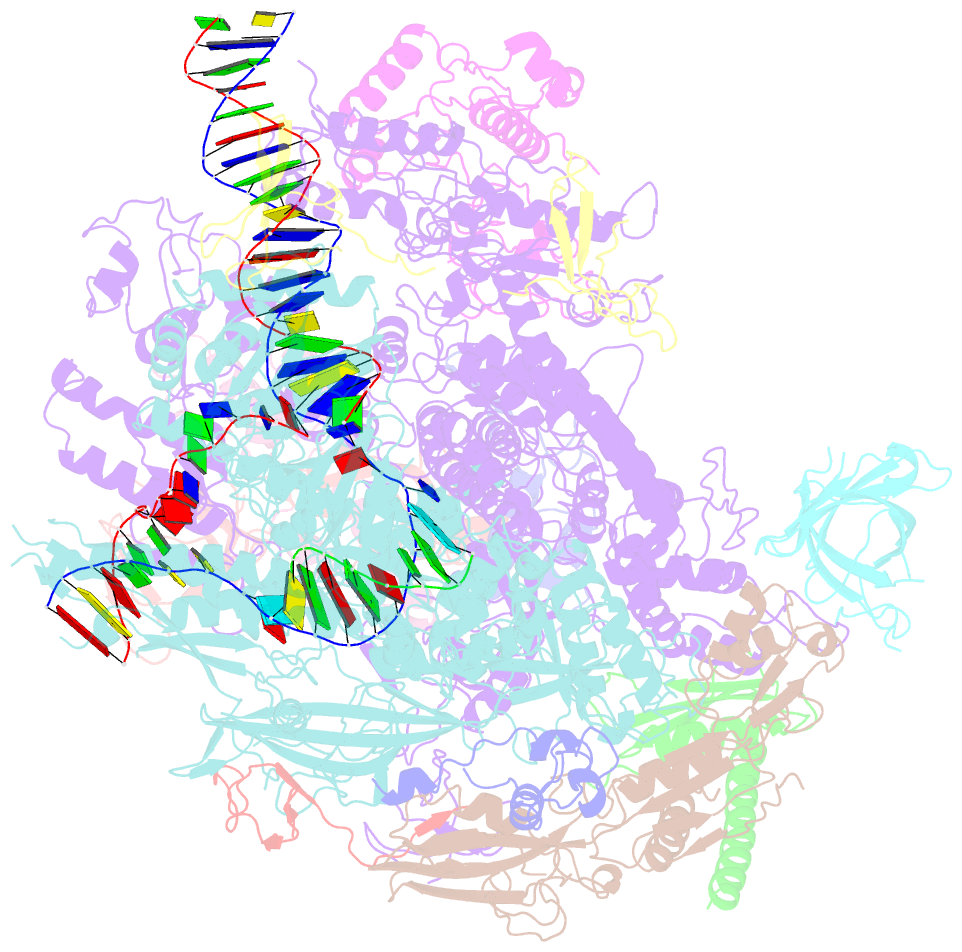
- A cryo-EM structure of ktf1-bound polymerase v transcription elongation complex
- Reference
- Zhang HW, Huang K, Gu ZX, Wu XX, Wang JW, Zhang Y (2023): "A cryo-EM structure of KTF1-bound polymerase V transcription elongation complex." Nat Commun, 14, 3118. doi: 10.1038/s41467-023-38619-x.
- Abstract
- De novo DNA methylation in plants relies on transcription of RNA polymerase V (Pol V) along with KTF1, which produce long non-coding RNAs for recruitment and assembly of the DNA methylation machinery. Here, we report a cryo-EM structure of the Pol V transcription elongation complex bound to KTF1. The structure reveals the conformation of the structural motifs in the active site of Pol V that accounts for its inferior RNA-extension ability. The structure also reveals structural features of Pol V that prevent it from interacting with the transcription factors of Pol II and Pol IV. The KOW5 domain of KTF1 binds near the RNA exit channel of Pol V providing a scaffold for the proposed recruitment of Argonaute proteins to initiate the assembly of the DNA methylation machinery. The structure provides insight into the Pol V transcription elongation process and the role of KTF1 during Pol V transcription-coupled DNA methylation.