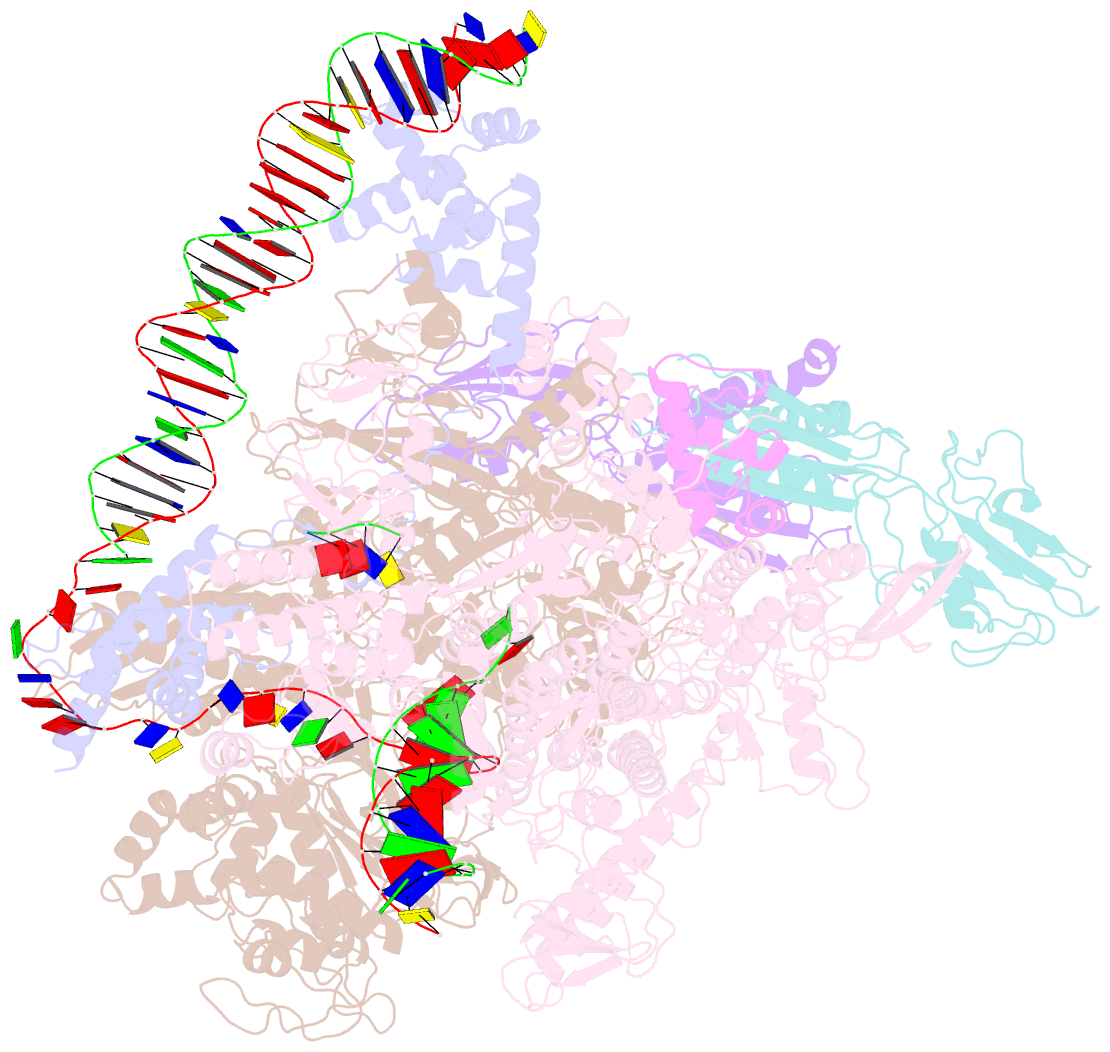
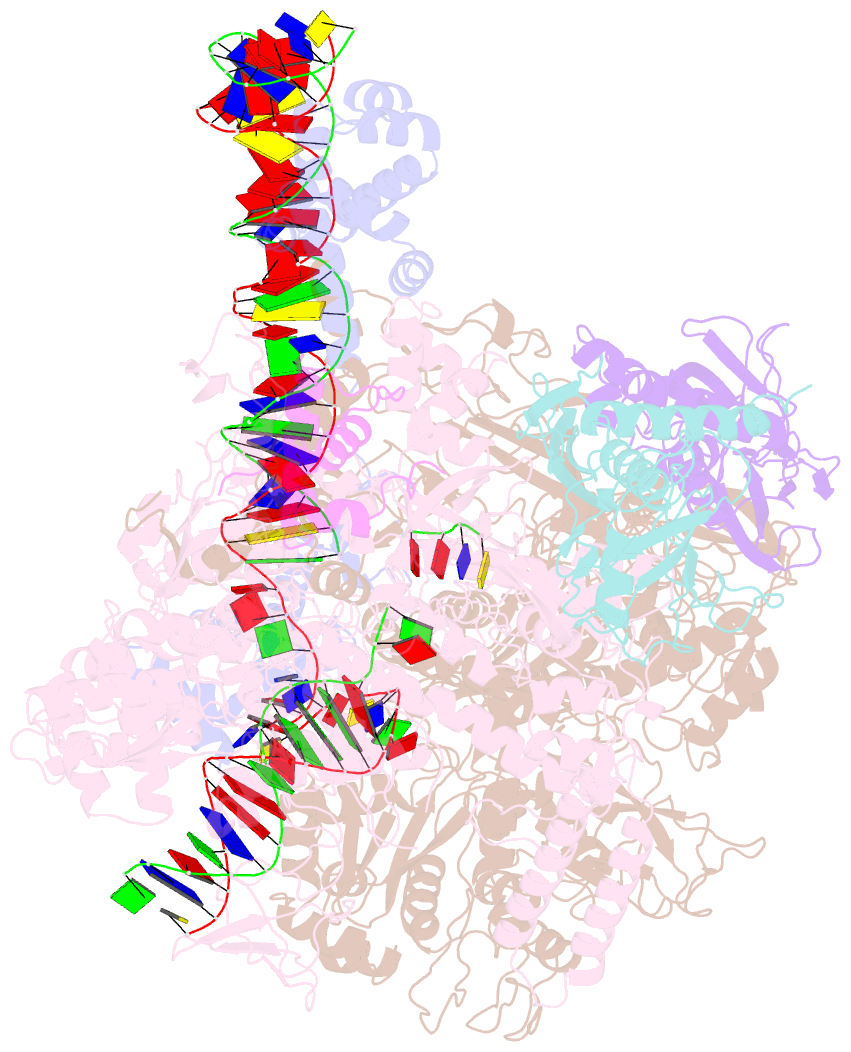
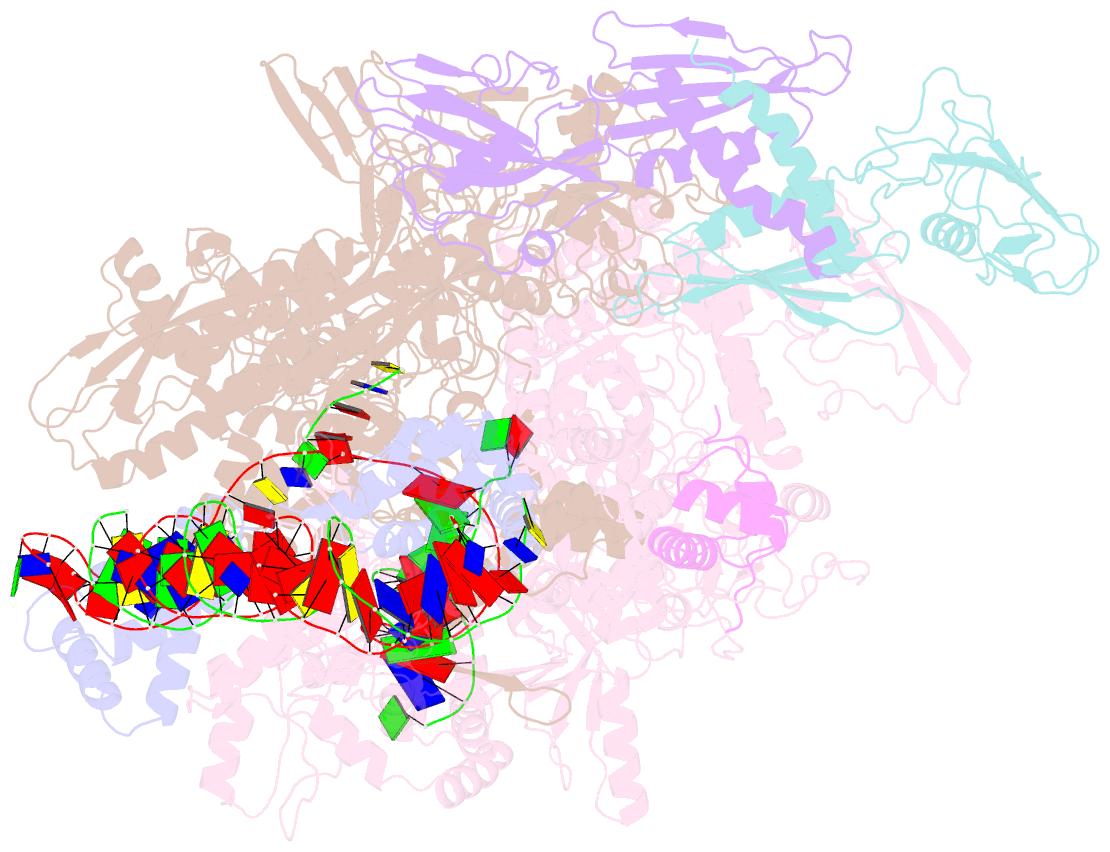
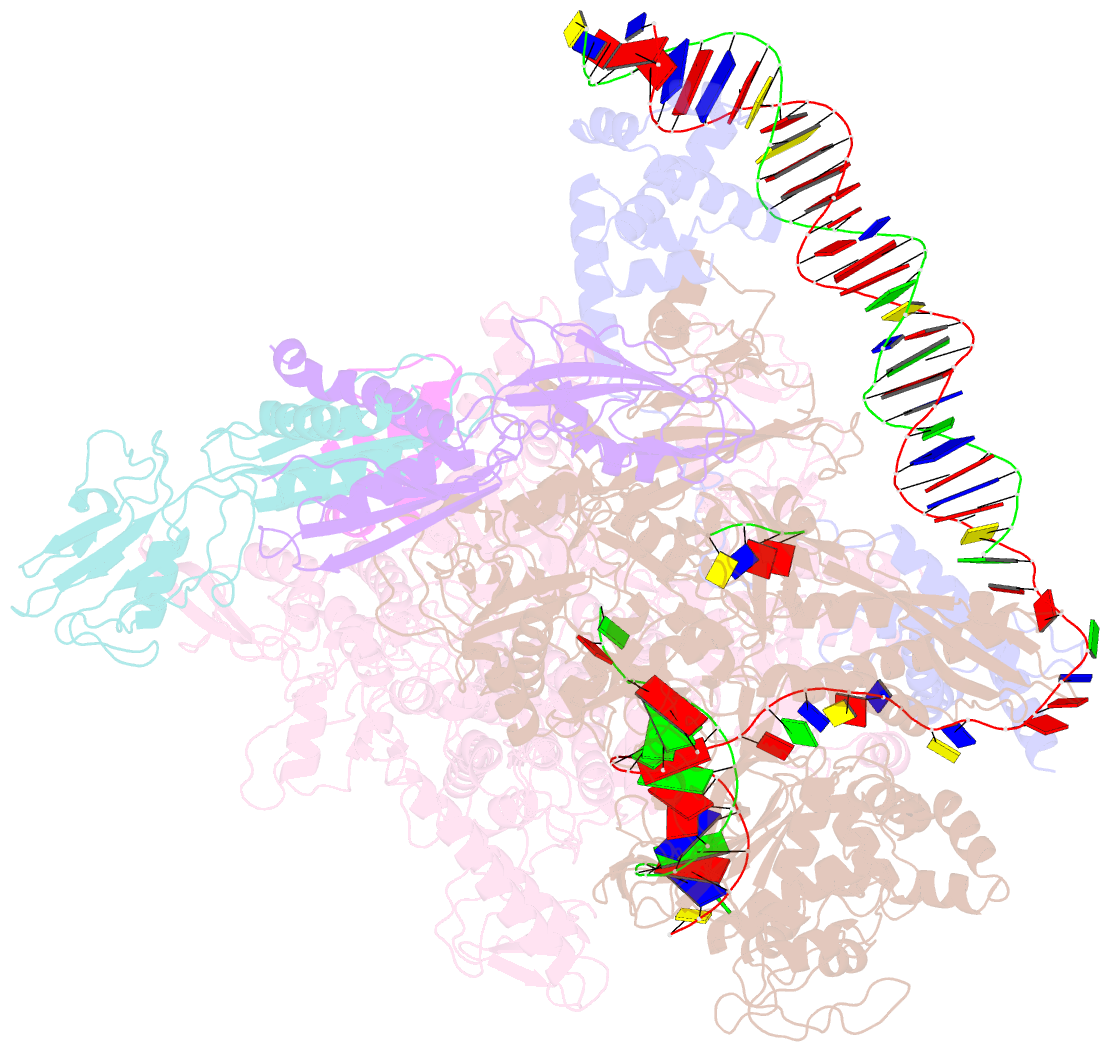
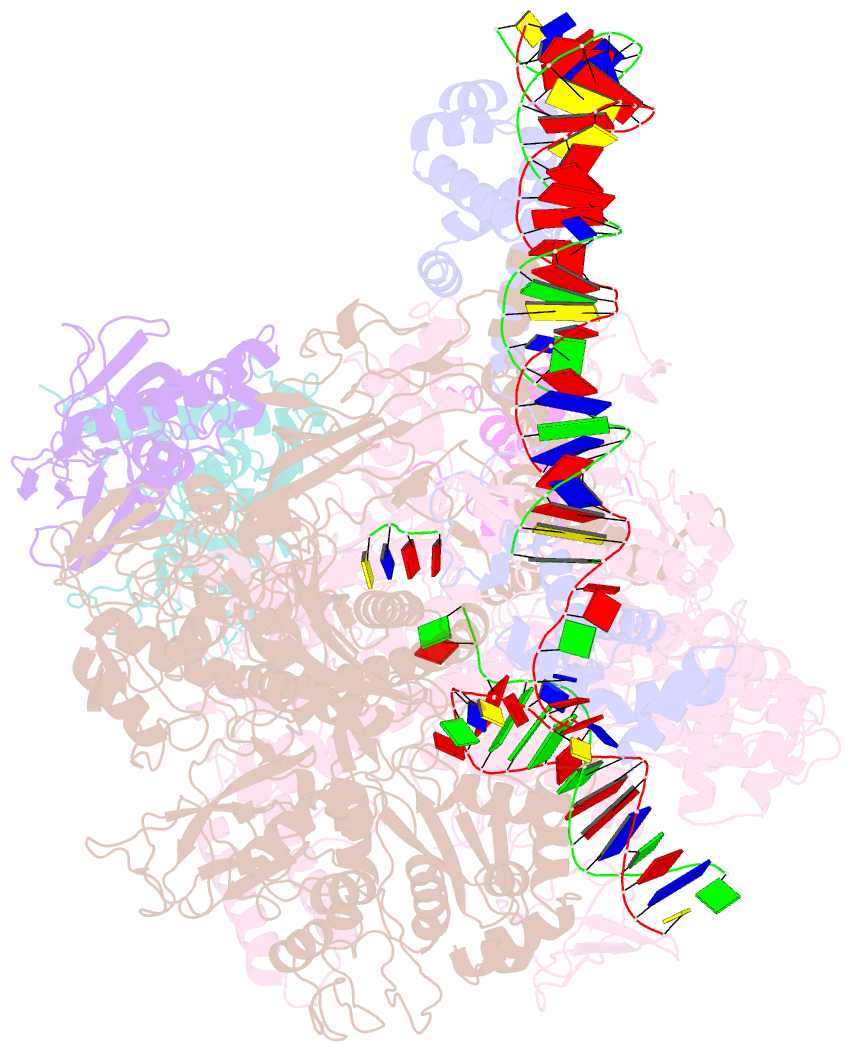
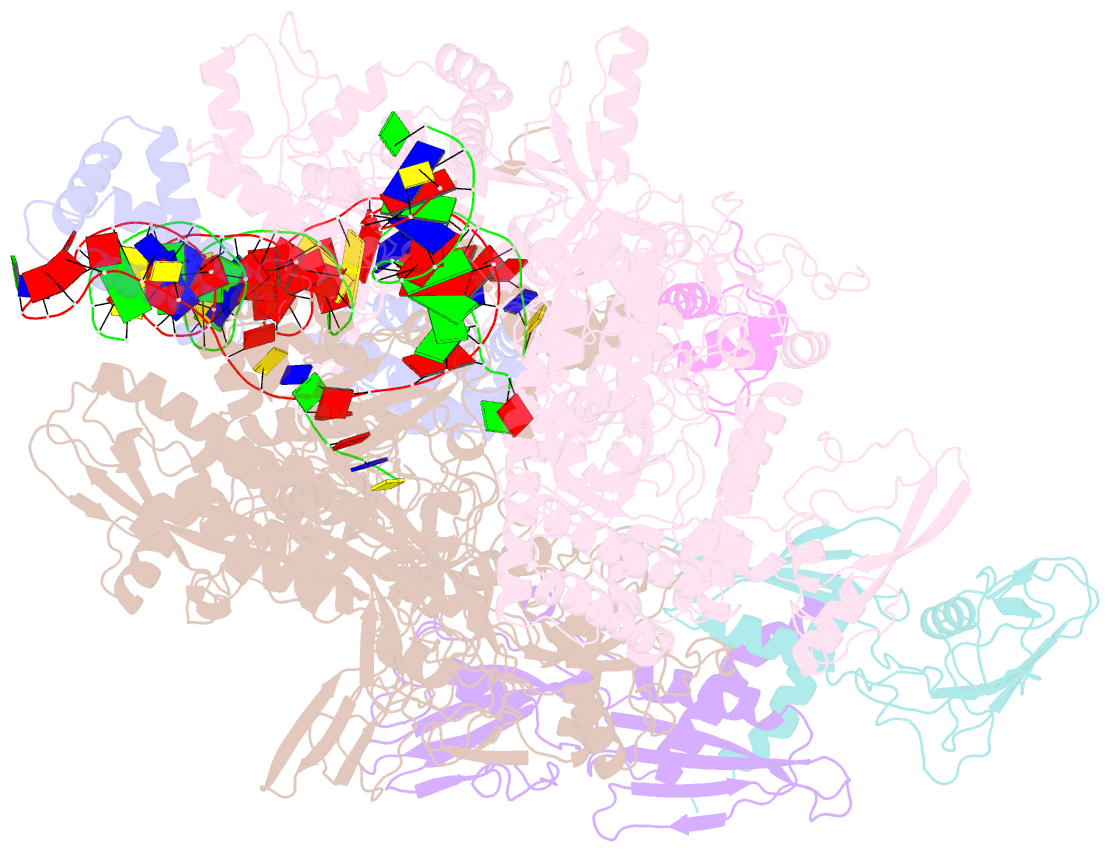
Summary information and primary citation
- PDB-id
- 8i23; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- cryo-EM (3.03 Å)
- Summary
- Clostridium thermocellum RNA polymerase transcription open complex with sigi1 and its promoter
- Reference
- Li J, Zhang H, Li D, Liu YJ, Bayer EA, Cui Q, Feng Y, Zhu P (2023): "Structure of the transcription open complex of distinct sigma I factors." Nat Commun, 14, 6455. doi: 10.1038/s41467-023-41796-4.
- Abstract
- Bacterial σI factors of the σ70-family are widespread in Bacilli and Clostridia and are involved in the heat shock response, iron metabolism, virulence, and carbohydrate sensing. A multiplicity of σI paralogues in some cellulolytic bacteria have been shown to be responsible for the regulation of the cellulosome, a multienzyme complex that mediates efficient cellulose degradation. Here, we report two structures at 3.0 Å and 3.3 Å of two transcription open complexes formed by two σI factors, SigI1 and SigI6, respectively, from the thermophilic, cellulolytic bacterium, Clostridium thermocellum. These structures reveal a unique, hitherto-unknown recognition mode of bacterial transcriptional promoters, both with respect to domain organization and binding to promoter DNA. The key characteristics that determine the specificities of the σI paralogues were further revealed by comparison of the two structures. Consequently, the σI factors represent a distinct set of the σ70-family σ factors, thus highlighting the diversity of bacterial transcription.