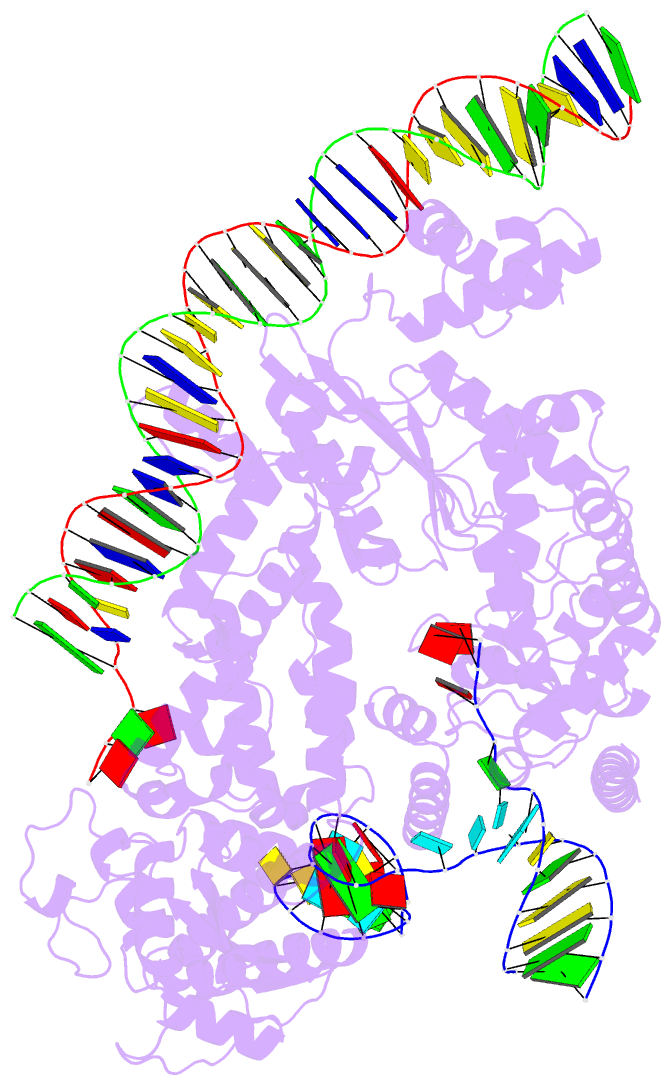
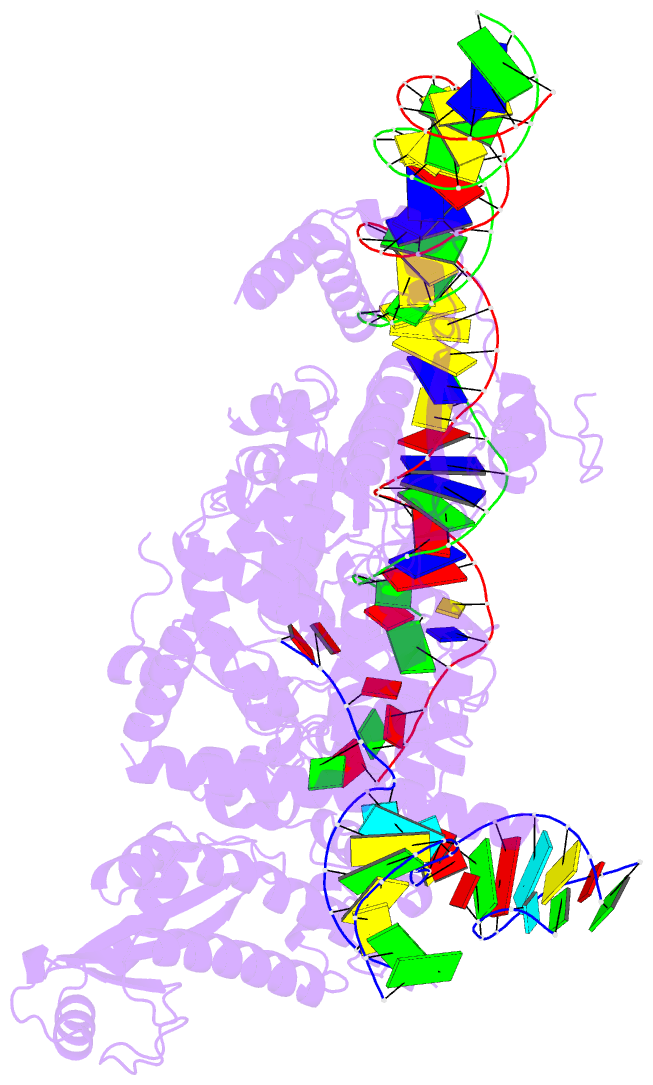
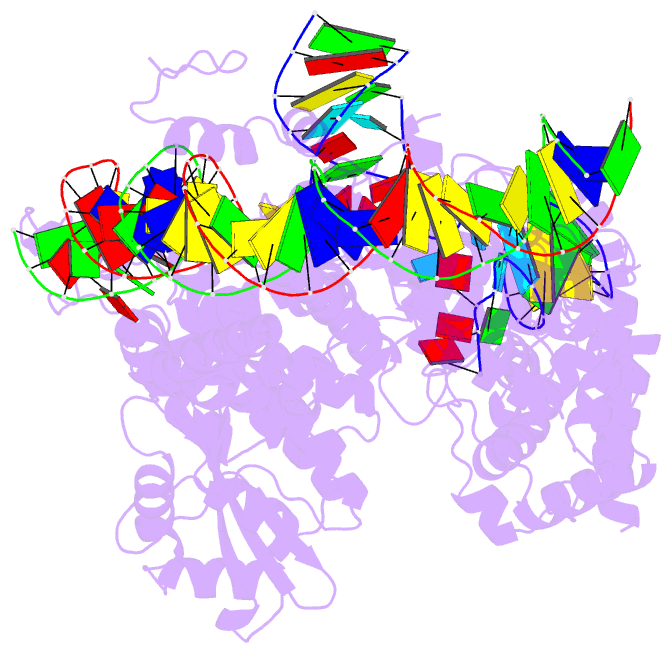
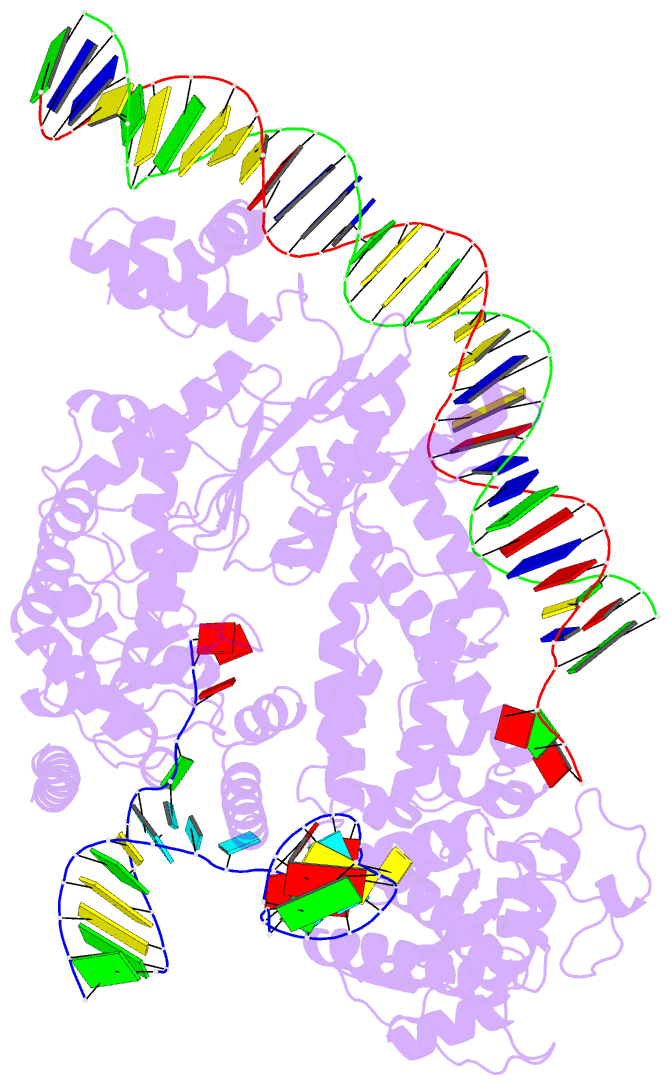
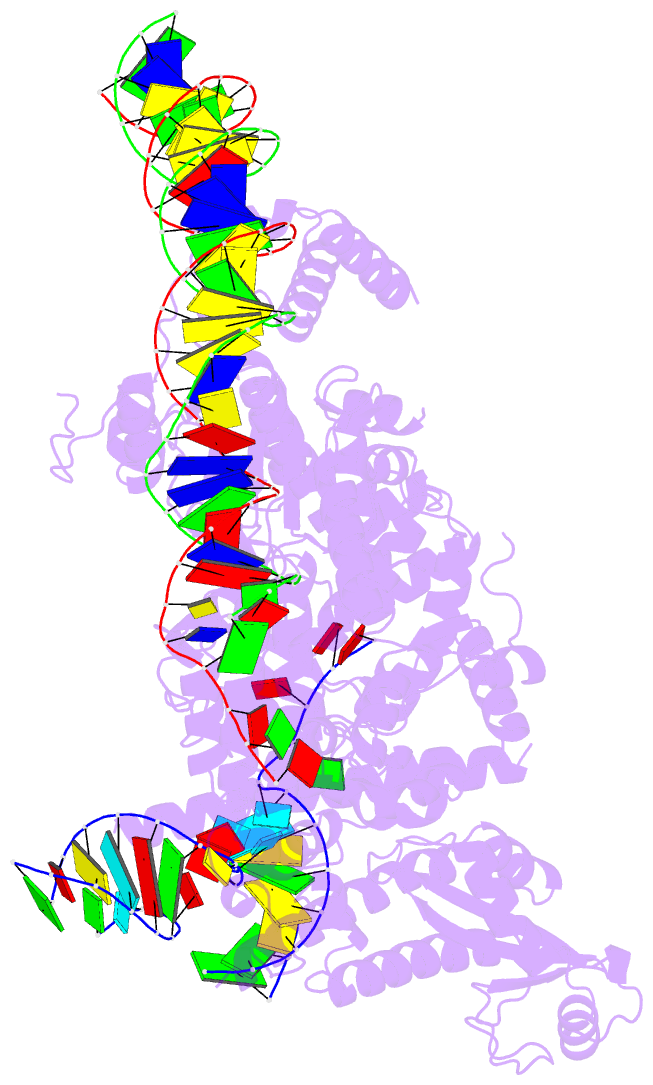
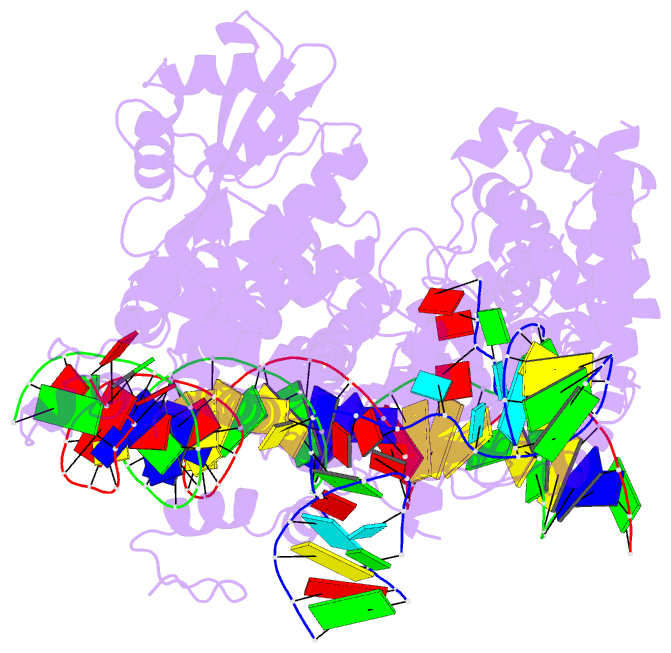
Summary information and primary citation
- PDB-id
- 8ibx; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA-DNA
- Method
- cryo-EM (3.74 Å)
- Summary
- Structure of r2 with 3'utr and DNA in unwinding state
- Reference
- Deng P, Tan SQ, Yang QY, Fu L, Wu Y, Zhu HZ, Sun L, Bao Z, Lin Y, Zhang QC, Wang H, Wang J, Liu JG (2023): "Structural RNA components supervise the sequential DNA cleavage in R2 retrotransposon." Cell, 186, 2865-2879.e20. doi: 10.1016/j.cell.2023.05.032.
- Abstract
- Retroelements are the widespread jumping elements considered as major drivers for genome evolution, which can also be repurposed as gene-editing tools. Here, we determine the cryo-EM structures of eukaryotic R2 retrotransposon with ribosomal DNA target and regulatory RNAs. Combined with biochemical and sequencing analysis, we reveal two essential DNA regions, Drr and Dcr, required for recognition and cleavage. The association of 3' regulatory RNA with R2 protein accelerates the first-strand cleavage, blocks the second-strand cleavage, and initiates the reverse transcription starting from the 3'-tail. Removing 3' regulatory RNA by reverse transcription allows the association of 5' regulatory RNA and initiates the second-strand cleavage. Taken together, our work explains the DNA recognition and RNA supervised sequential retrotransposition mechanisms by R2 machinery, providing insights into the retrotransposon and application reprogramming.