Summary information and primary citation
- PDB-id
- 8igs; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- cryo-EM (3.4 Å)
- Summary
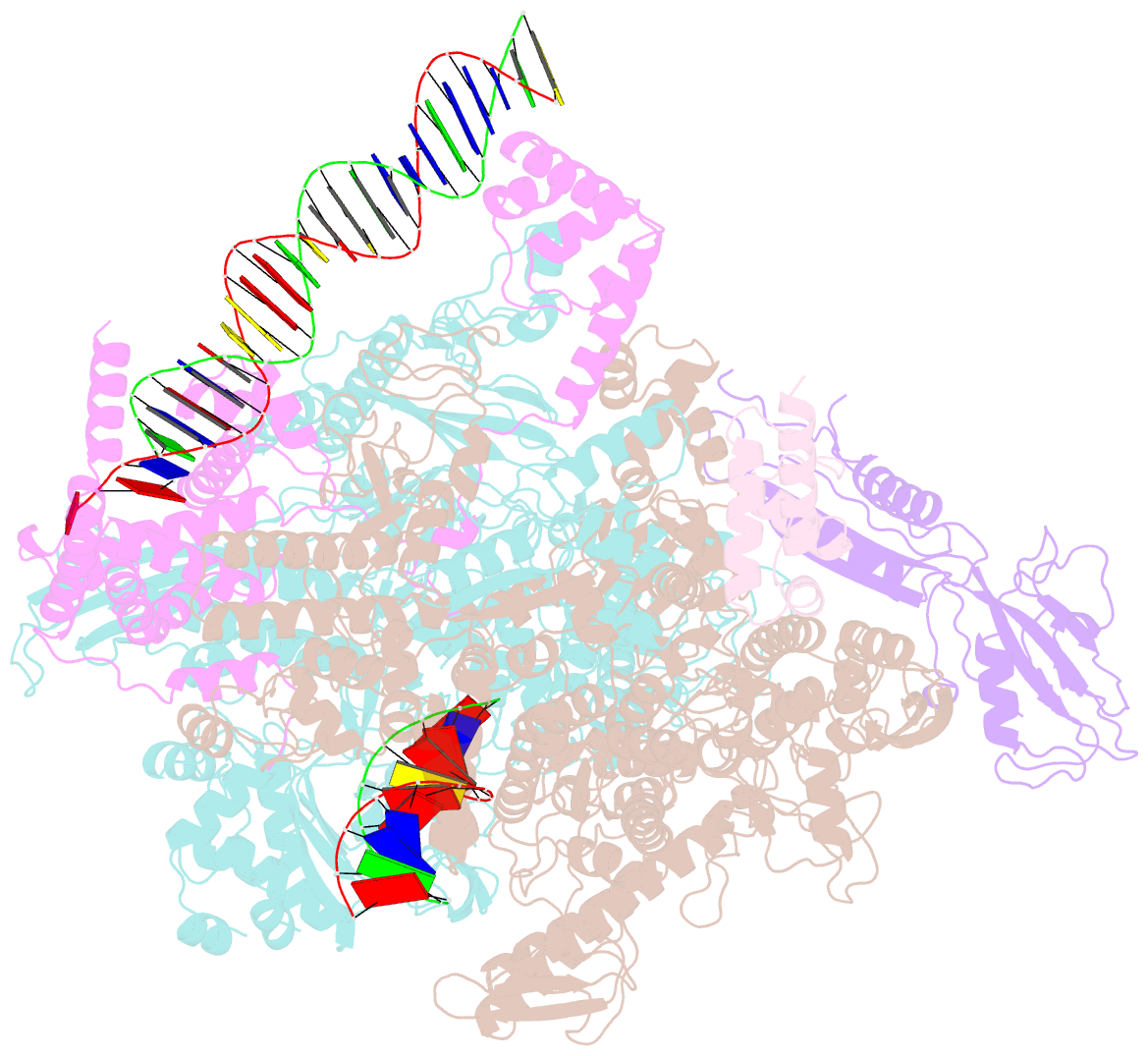
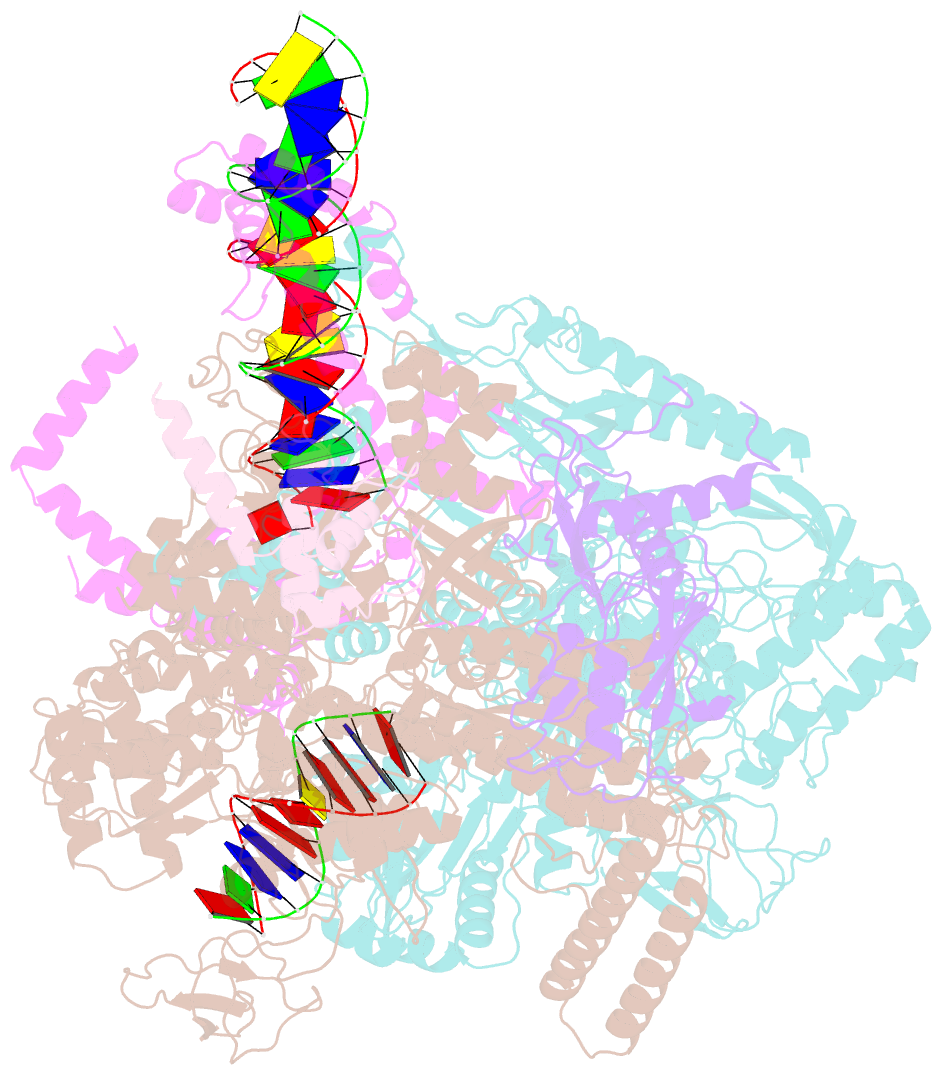
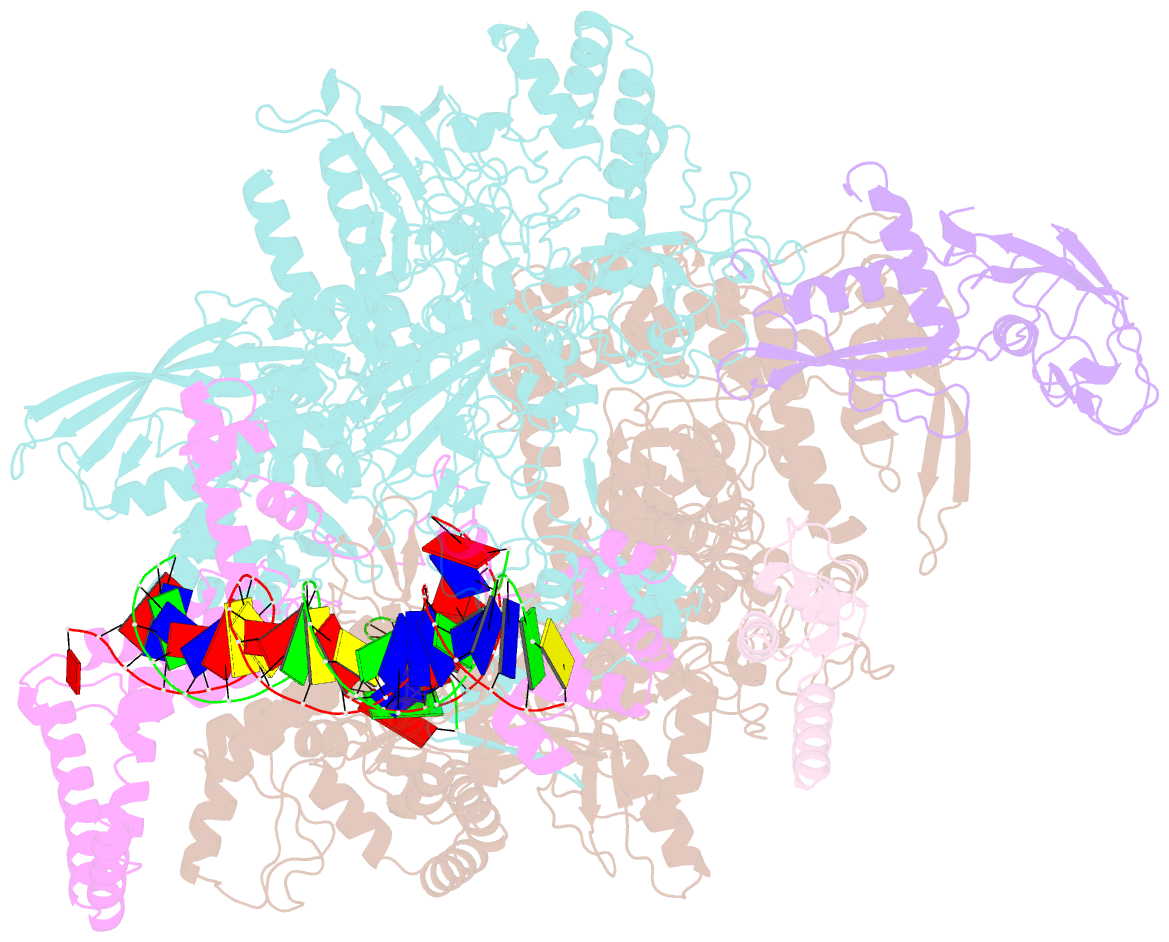
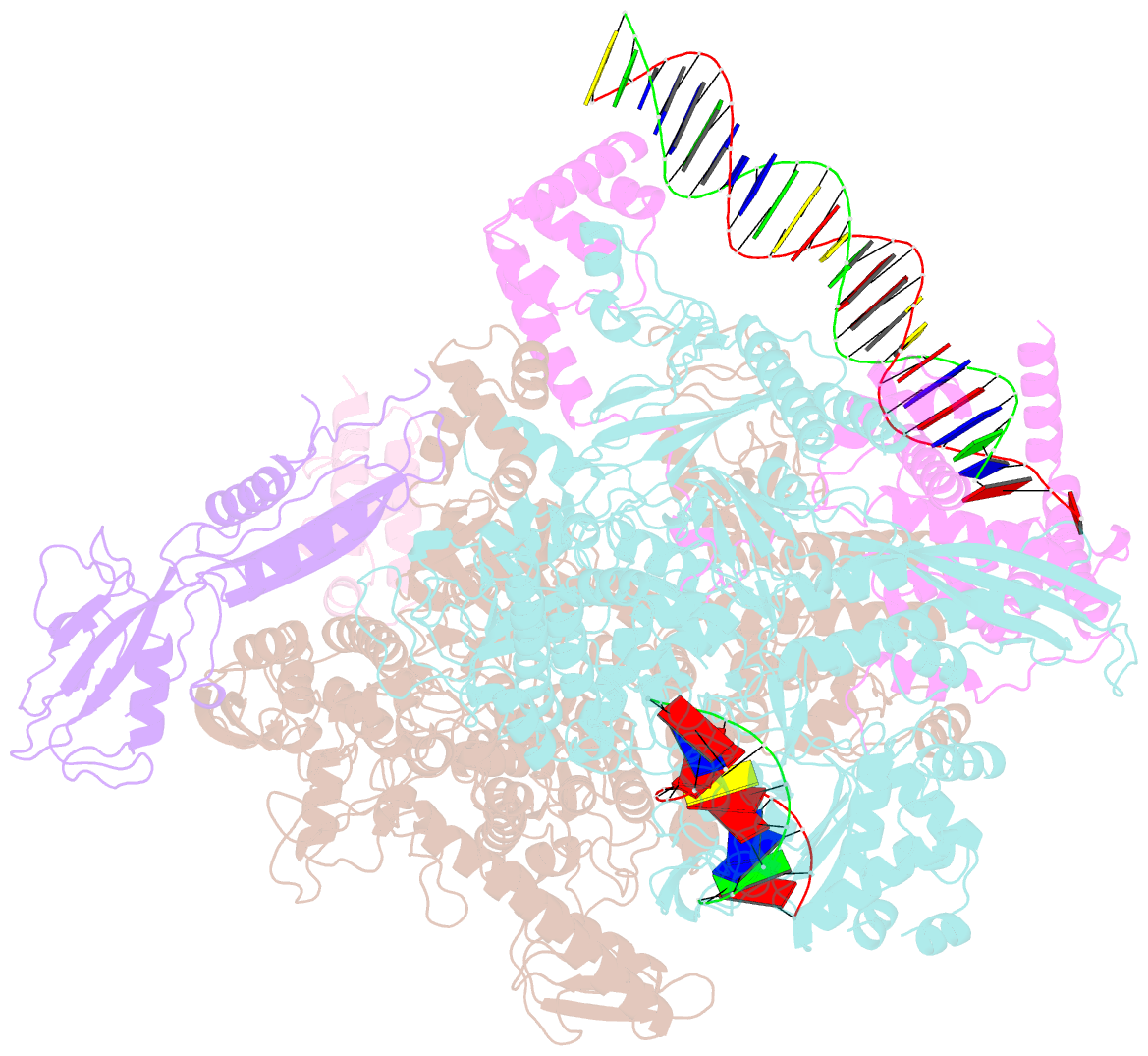
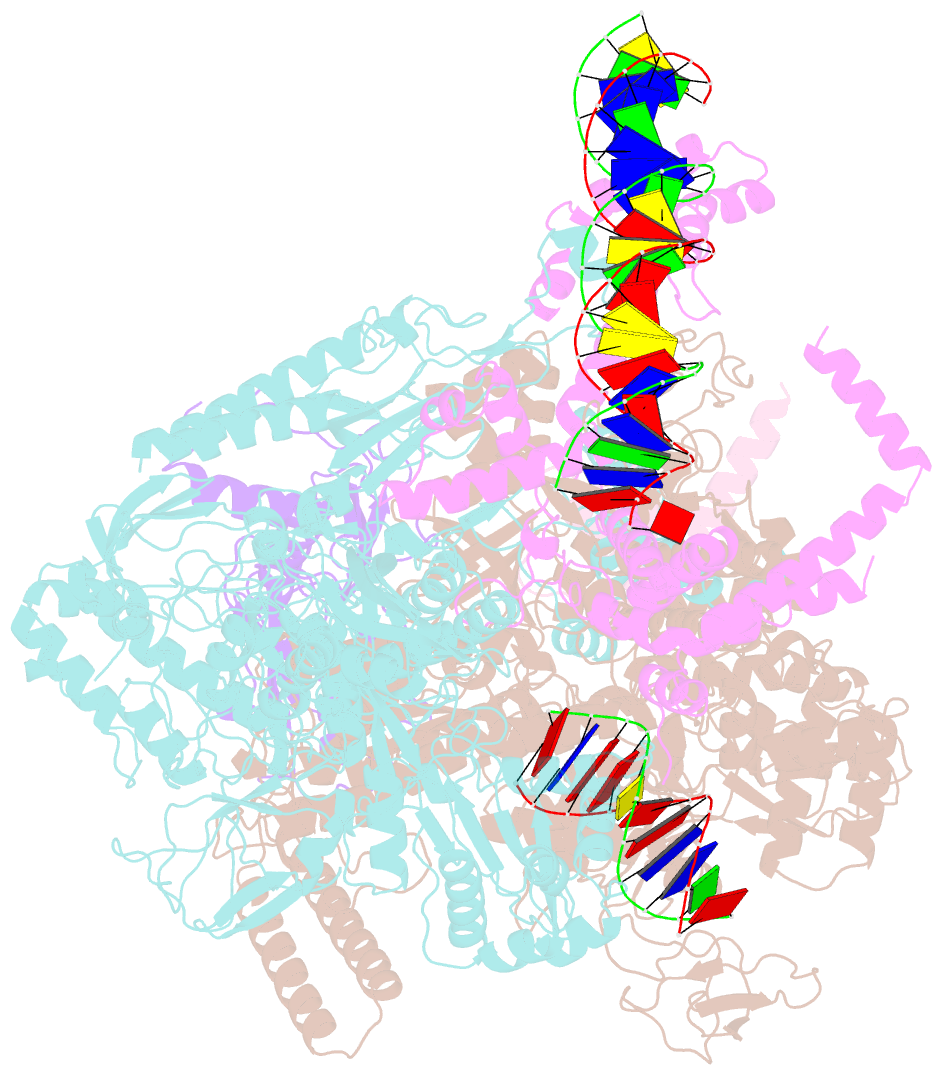
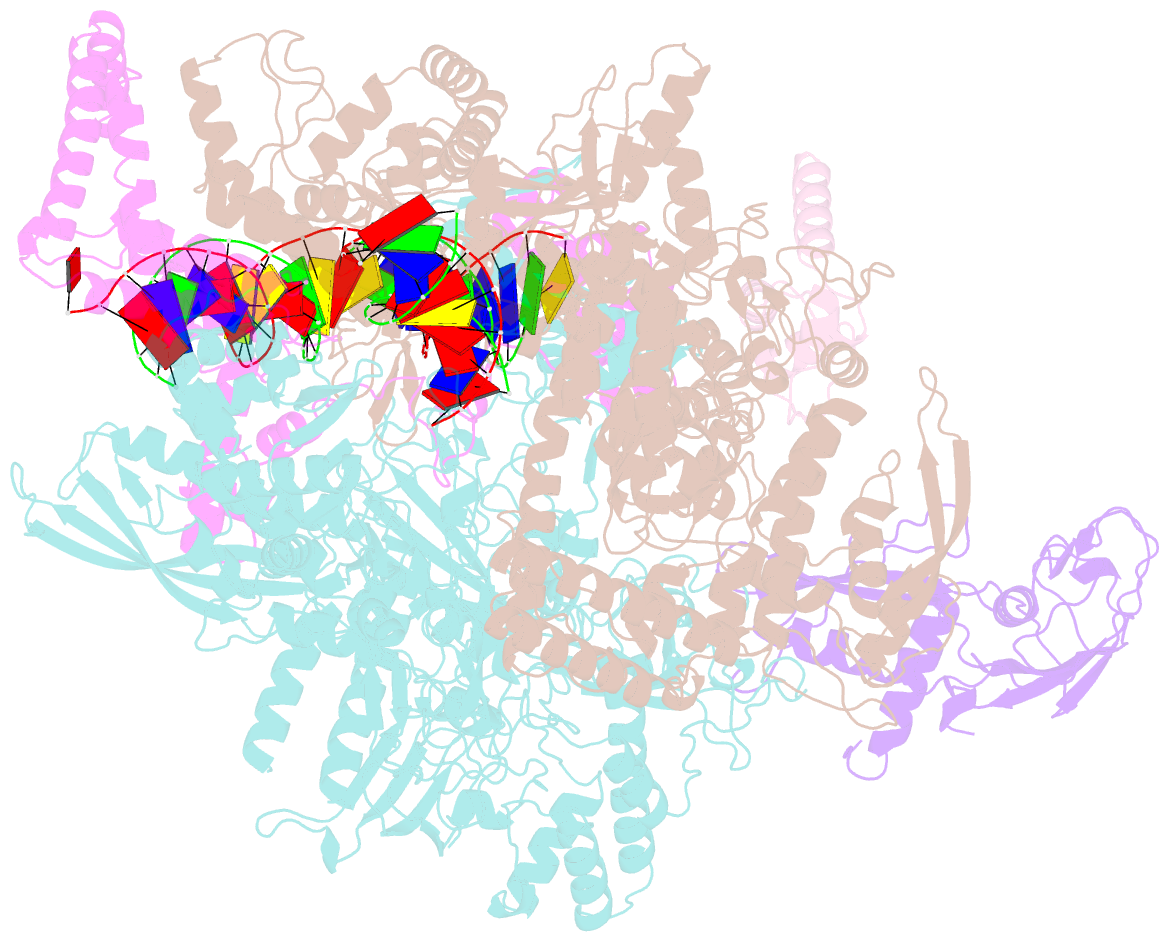
- cryo-EM structure of rnap-promoter open complex at lambda promoter pre
- Reference
- Zhao M, Gao B, Wen A, Feng Y, Lu YQ (2023): "Structural basis of lambda CII-dependent transcription activation." Structure, 31, 968. doi: 10.1016/j.str.2023.05.008.
- Abstract
- The CII protein of bacteriophage λ activates transcription from the phage promoters PRE, PI, and PAQ by binding to two direct repeats that straddle the promoter -35 element. Although genetic, biochemical, and structural studies have elucidated many aspects of λCII-mediated transcription activation, no precise structure of the transcription machinery in the process is available. Here, we report a 3.1-Å cryo-electron microscopy (cryo-EM) structure of an intact λCII-dependent transcription activation complex (TAC-λCII), which comprises λCII, E. coli RNAP-σ70 holoenzyme, and the phage promoter PRE. The structure reveals the interactions between λCII and the direct repeats responsible for promoter specificity and the interactions between λCII and RNAP α subunit C-terminal domain responsible for transcription activation. We also determined a 3.4-Å cryo-EM structure of an RNAP-promoter open complex (RPo-PRE) from the same dataset. Structural comparison between TAC-λCII and RPo-PRE provides new insights into λCII-dependent transcription activation.