Summary information and primary citation
- PDB-id
- 8ipm; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ribosomal protein
- Method
- X-ray (3.1 Å)
- Summary
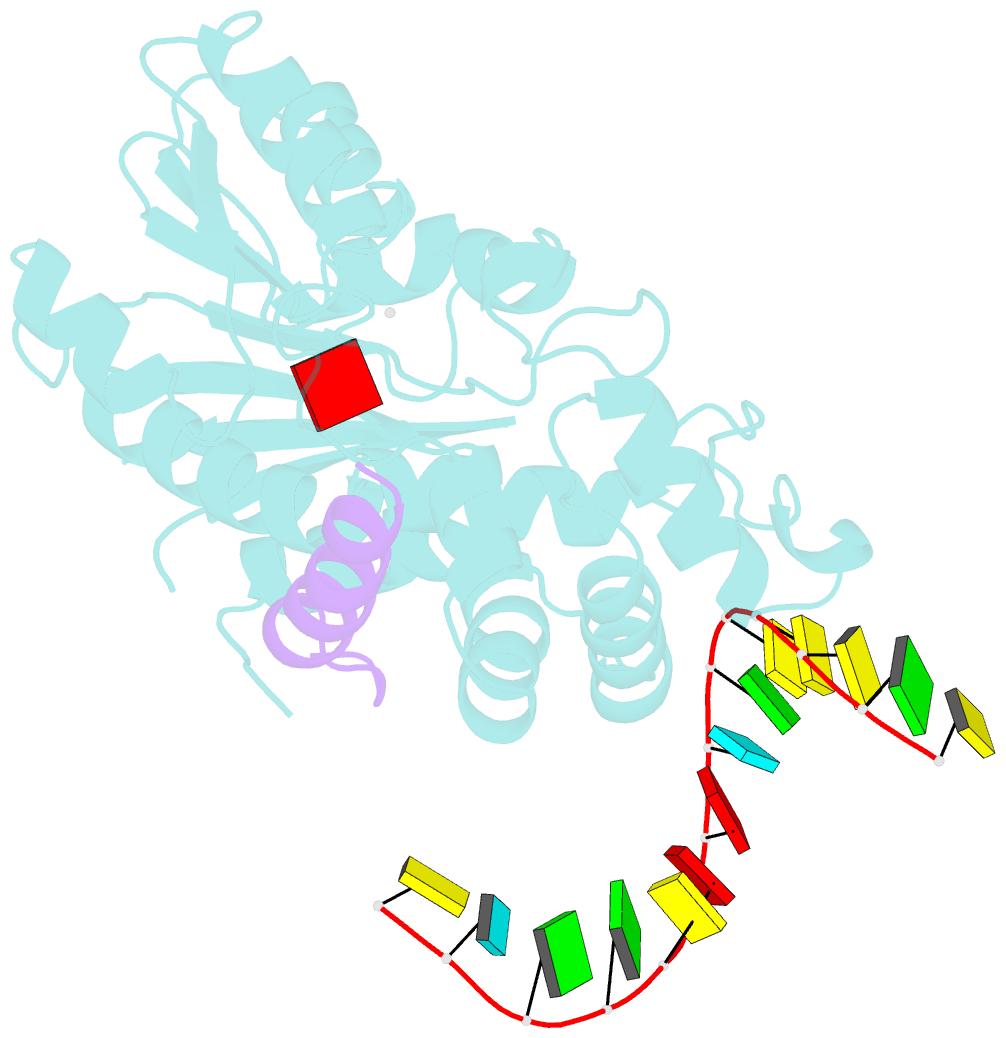
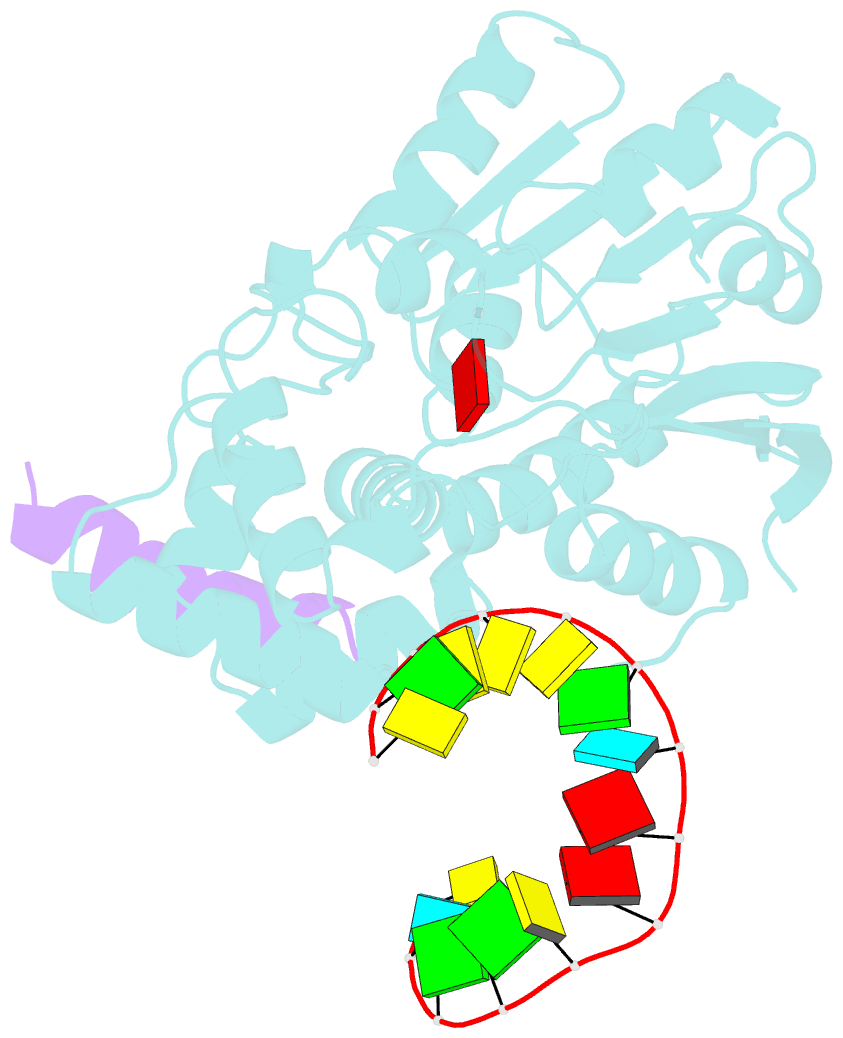
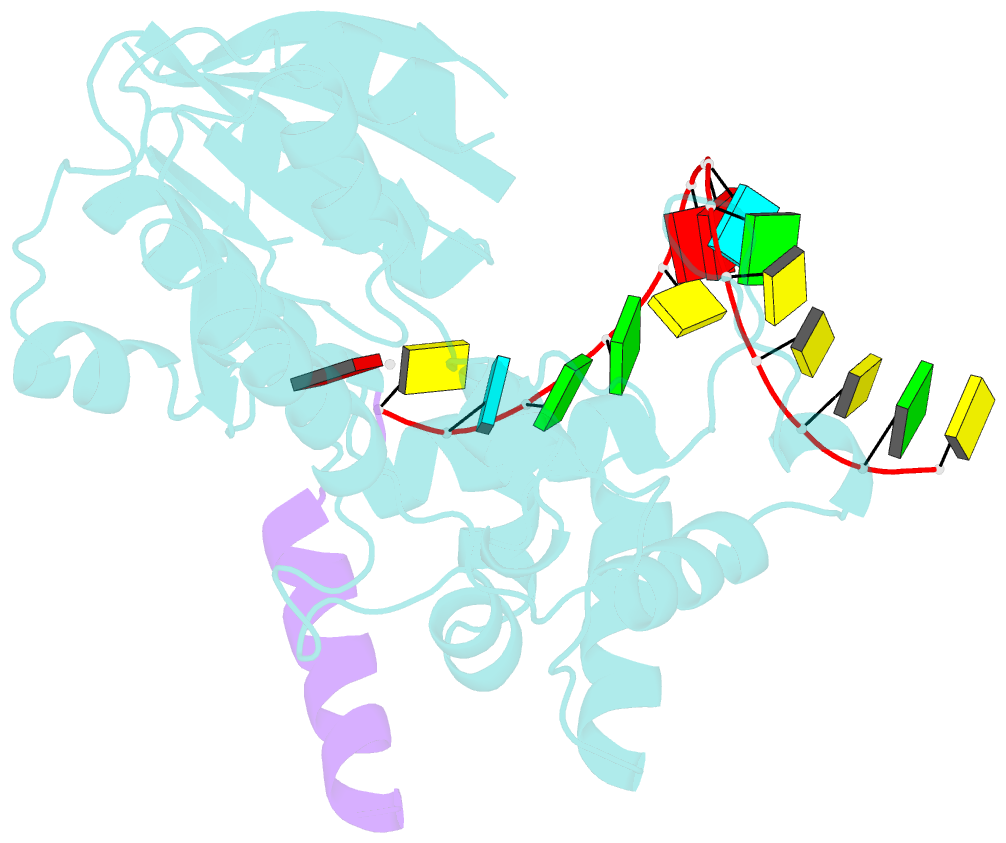
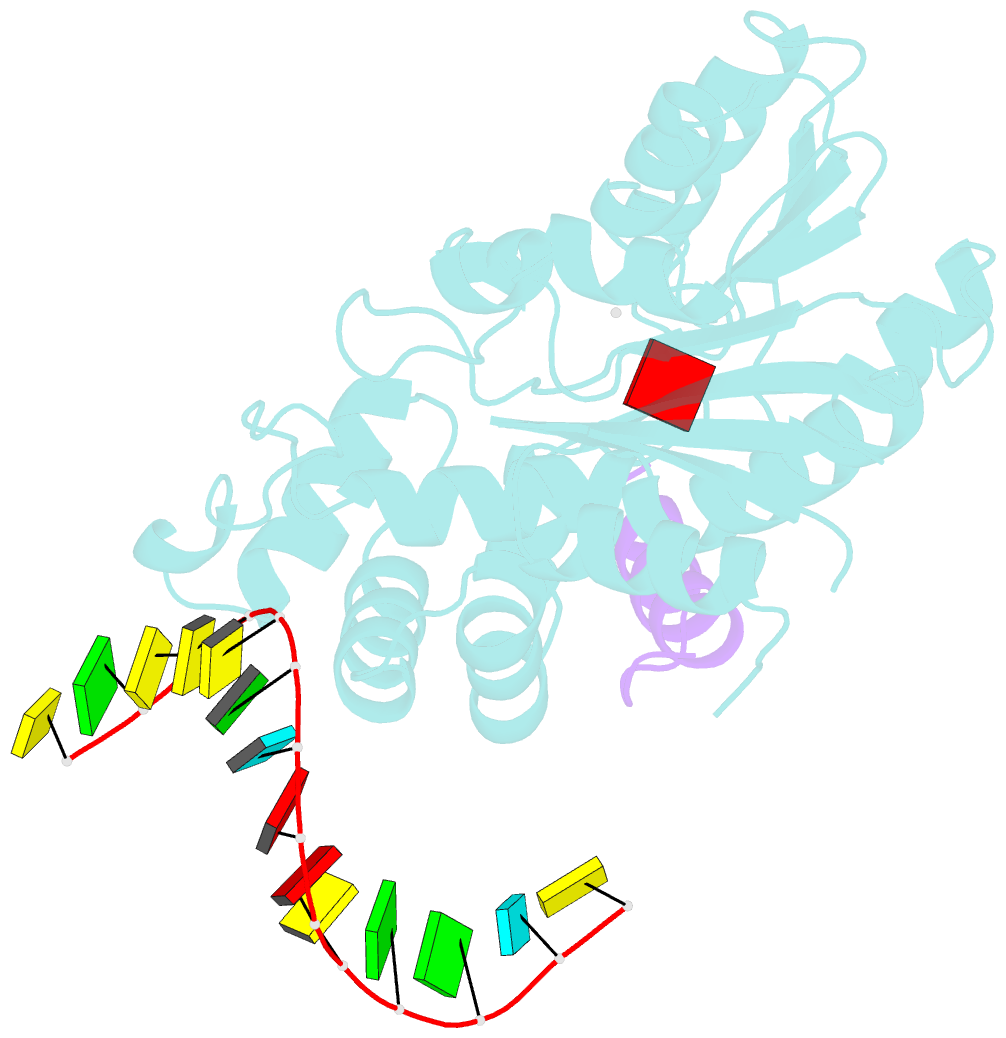
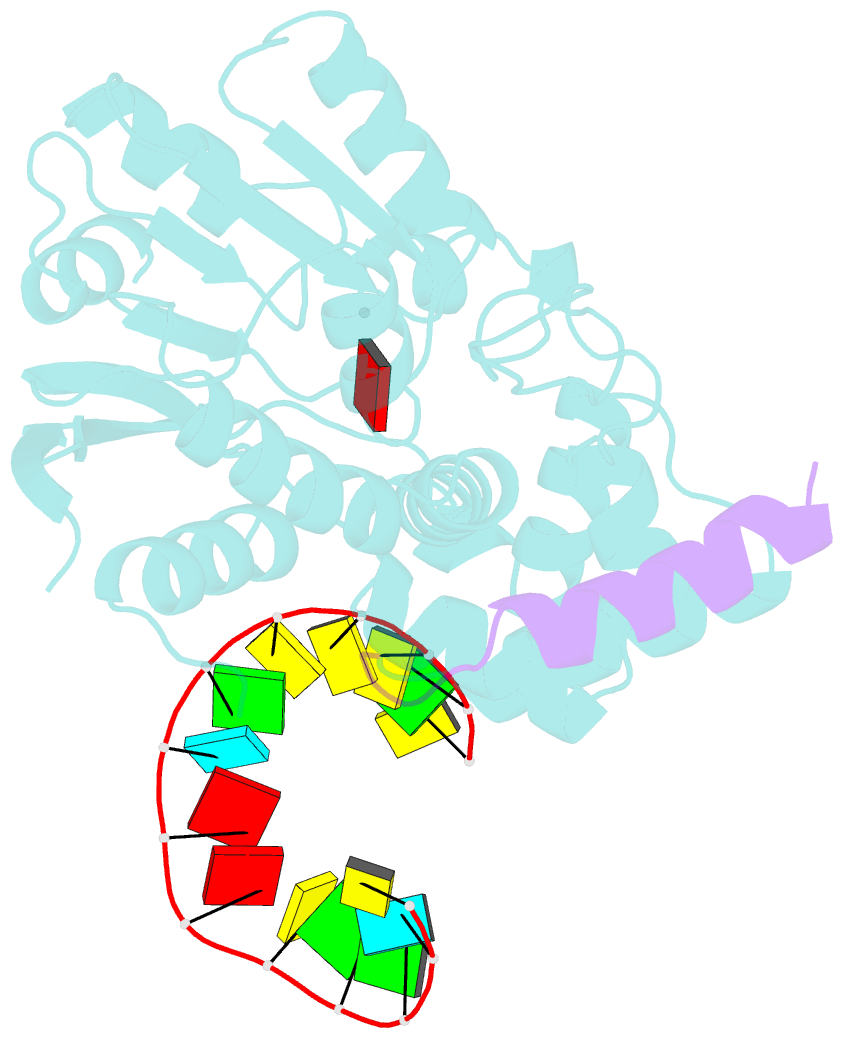
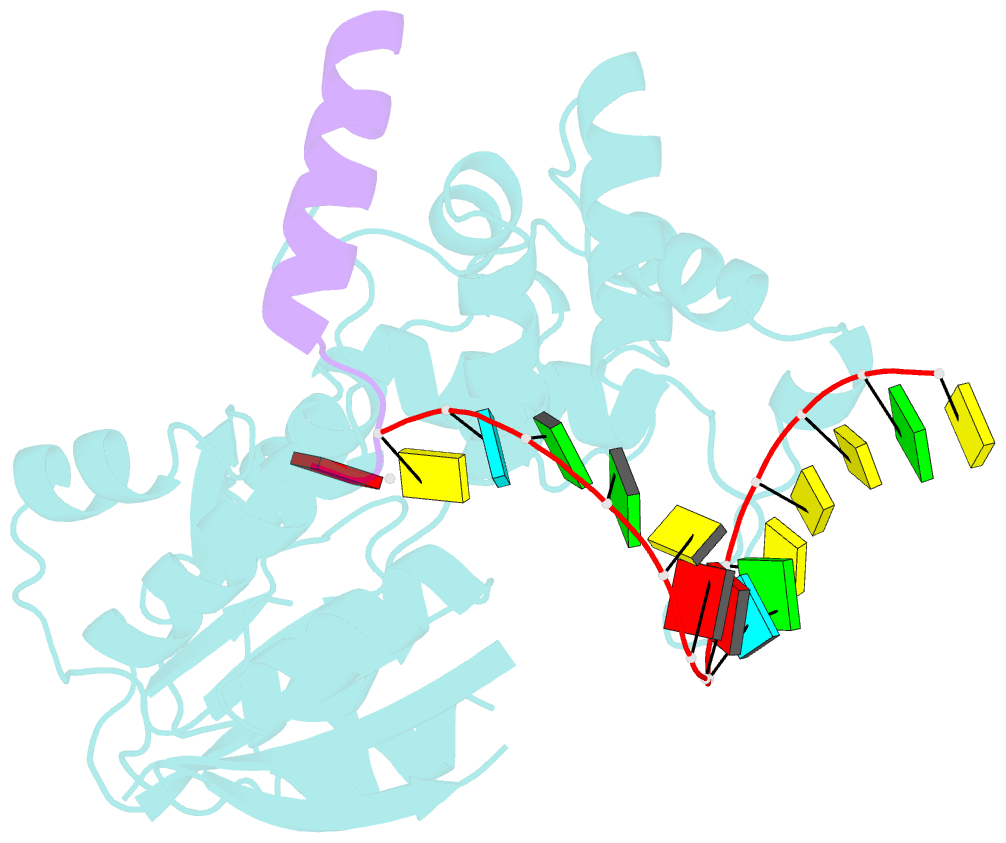
- The structure of human mitochondrial methyltransferase mettl15 with h44_rna, rbfa and sam
- Reference
- Lv M, Zhou W, Hao Y, Li F, Zhang H, Yao X, Shi Y, Zhang L (2024): "Structural insights into the specific recognition of mitochondrial ribosome-binding factor hsRBFA and 12 S rRNA by methyltransferase METTL15." Cell Discov, 10, 11. doi: 10.1038/s41421-023-00634-z.
- Abstract
- Mitochondrial rRNA modifications are essential for mitoribosome assembly and its proper function. The m4C methyltransferase METTL15 maintains mitochondrial homeostasis by catalyzing m4C839 located in 12 S rRNA helix 44 (h44). This modification is essential to fine-tuning the ribosomal decoding center and increasing decoding fidelity according to studies of a conserved site in Escherichia coli. Here, we reported a series of crystal structures of human METTL15-hsRBFA-h44-SAM analog, METTL15-hsRBFA-SAM, METTL15-SAM and apo METTL15. The structures presented specific interactions of METTL15 with different substrates and revealed that hsRBFA recruits METTL15 to mitochondrial small subunit for further modification instead of 12 S rRNA. Finally, we found that METTL15 deficiency caused increased reactive oxygen species, decreased membrane potential and altered cellular metabolic state. Knocking down METTL15 caused an elevated lactate secretion and increased levels of histone H4K12-lactylation and H3K9-lactylation. METTL15 might be a suitable model to study the regulation between mitochondrial metabolism and histone lactylation.