Summary information and primary citation
- PDB-id
- 8ipp; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (2.007 Å)
- Summary
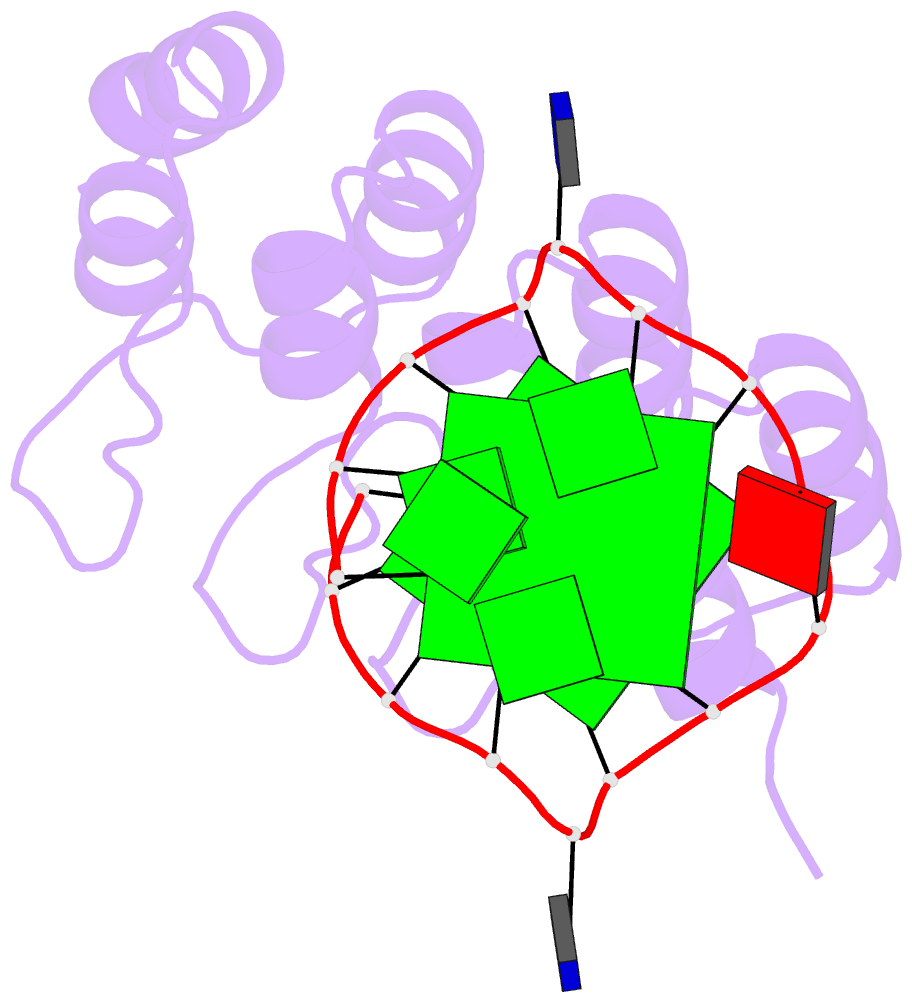
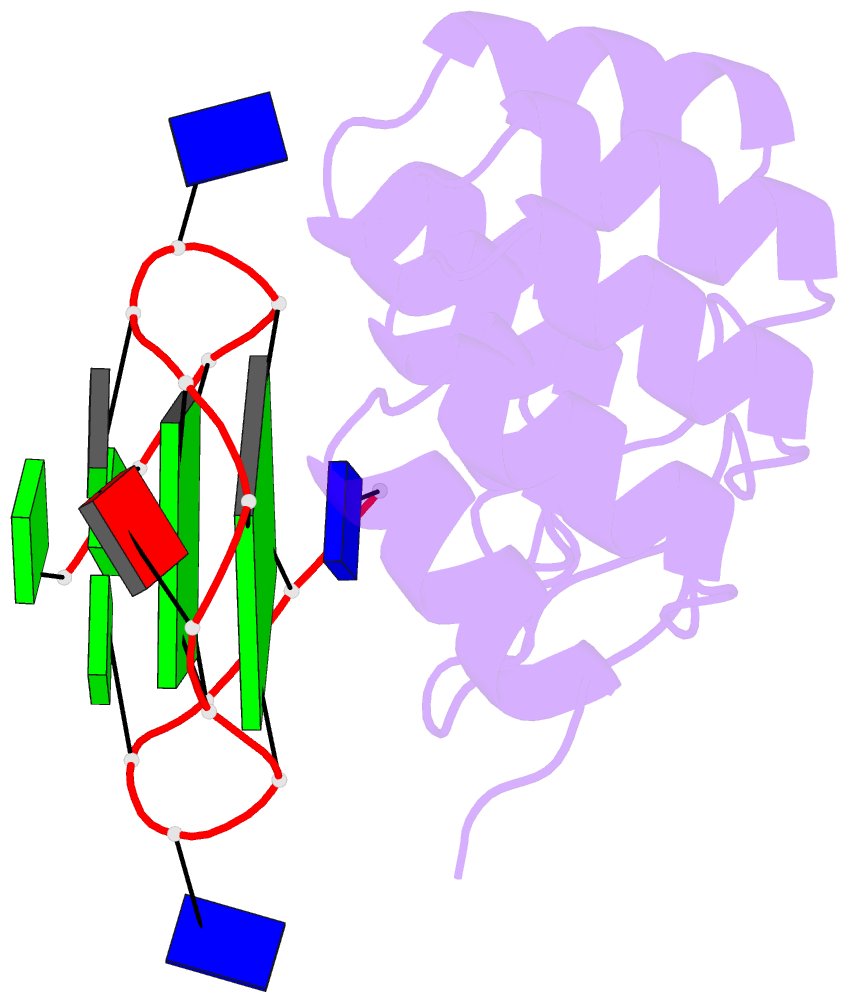
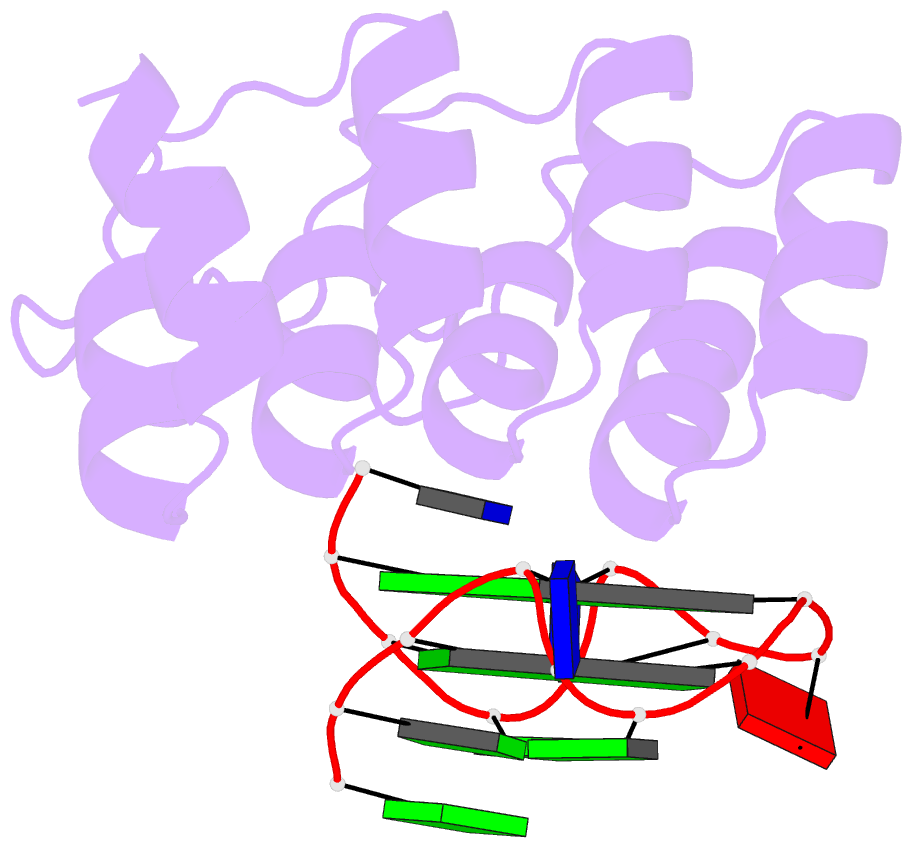
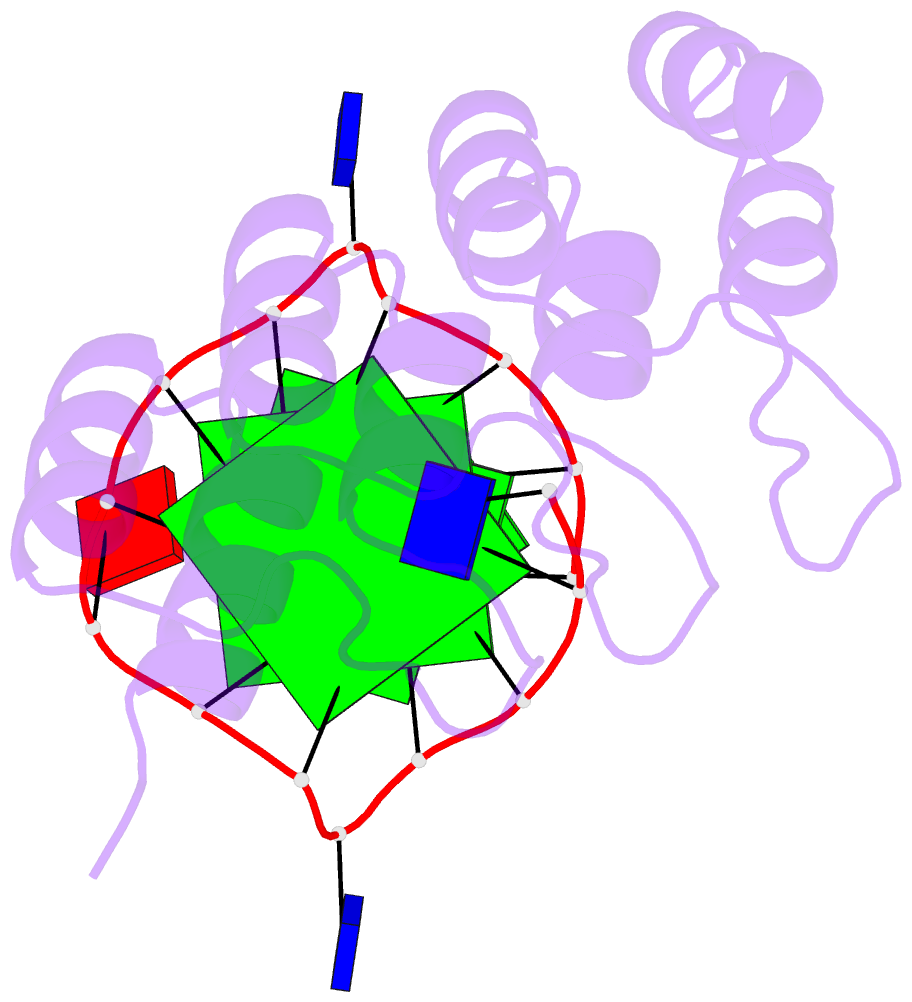
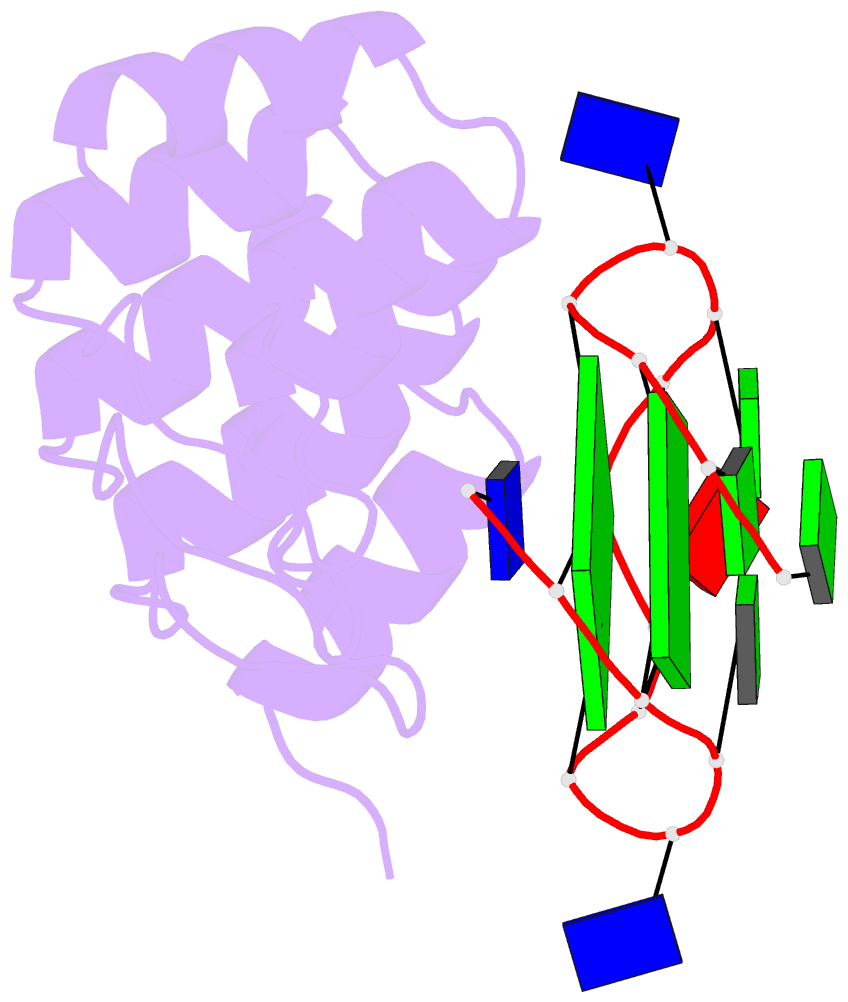
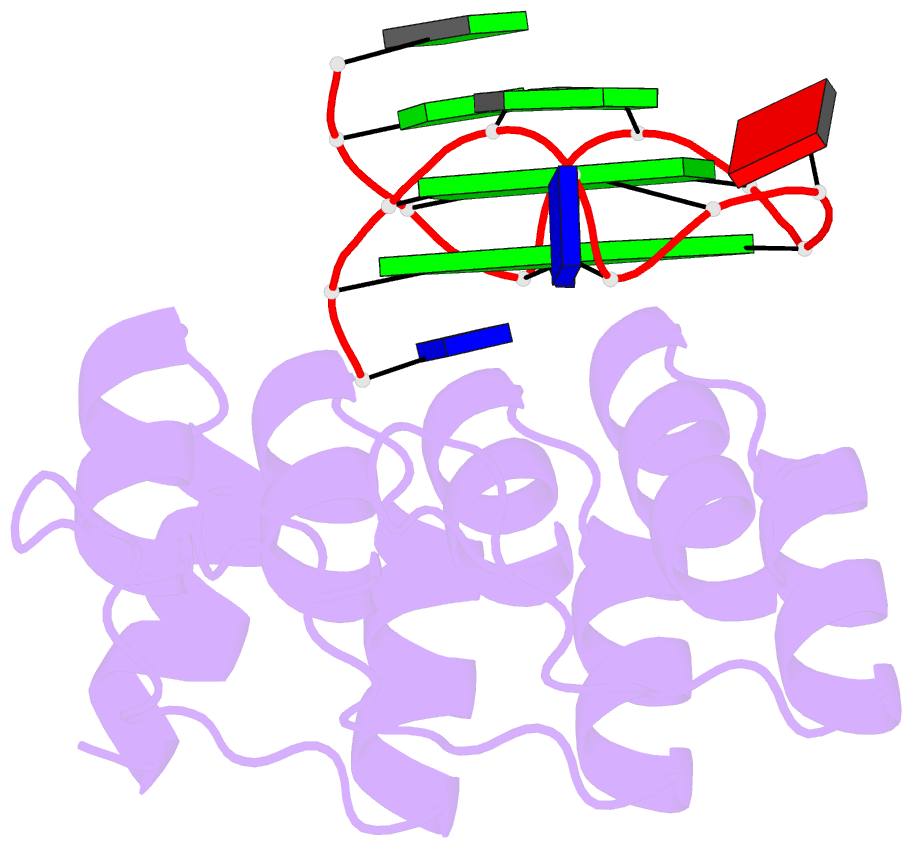
- Crystal structure of the complex between an ankyrin and a parallel g-quadruplex
- Reference
- Ngo KH, Liew CW, Heddi B, Phan AT (2024): "Structural Basis for Parallel G-Quadruplex Recognition by an Ankyrin Protein." J.Am.Chem.Soc., 146, 13709-13713. doi: 10.1021/jacs.4c01971.
- Abstract
- G-Quadruplex (G4) structures formed by guanine-rich DNA and RNA sequences are implicated in various biological processes. Understanding the mechanisms by which proteins recognize G4 structures is crucial for elucidating their functional roles. Here we present the X-ray crystal structure of an ankyrin protein bound to a parallel G4 structure. Our findings reveal a new specific recognition mode in which a bundle of α-helices and loops of the ankyrin form a flat surface to stack on the G-tetrad core. The protein employs a combination of hydrogen bonds and hydrophobic contacts to interact with the G4, and electrostatic interaction is used to enhance the binding affinity. This binding mechanism provides valuable insights into understanding G4 recognition by proteins.