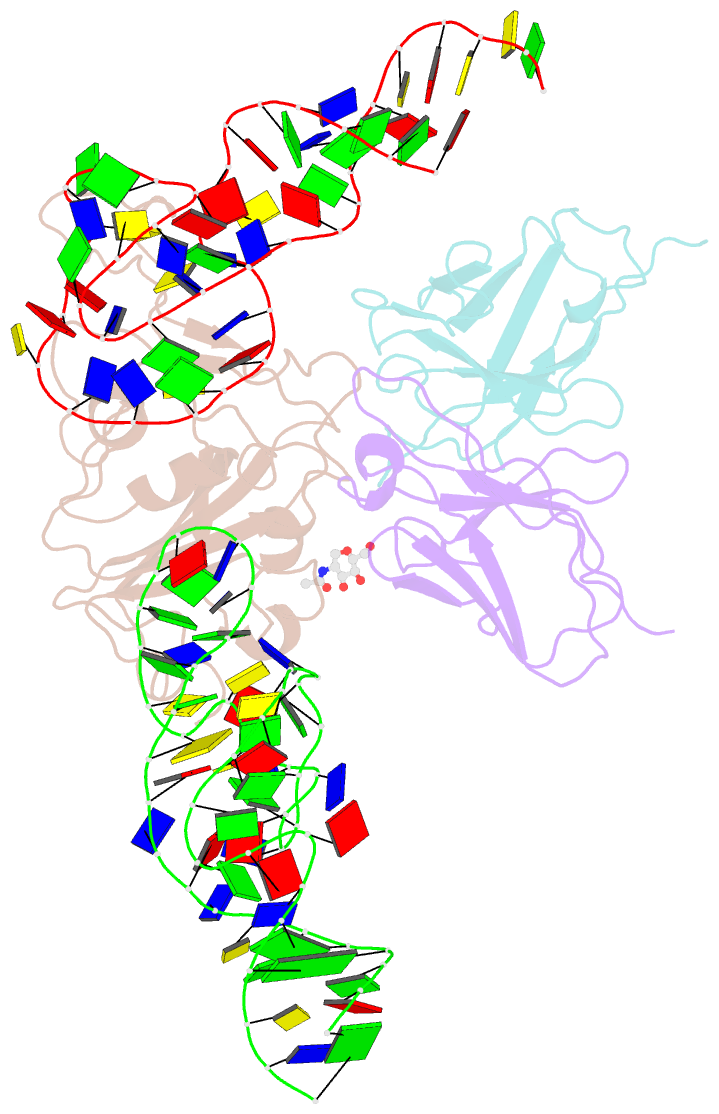
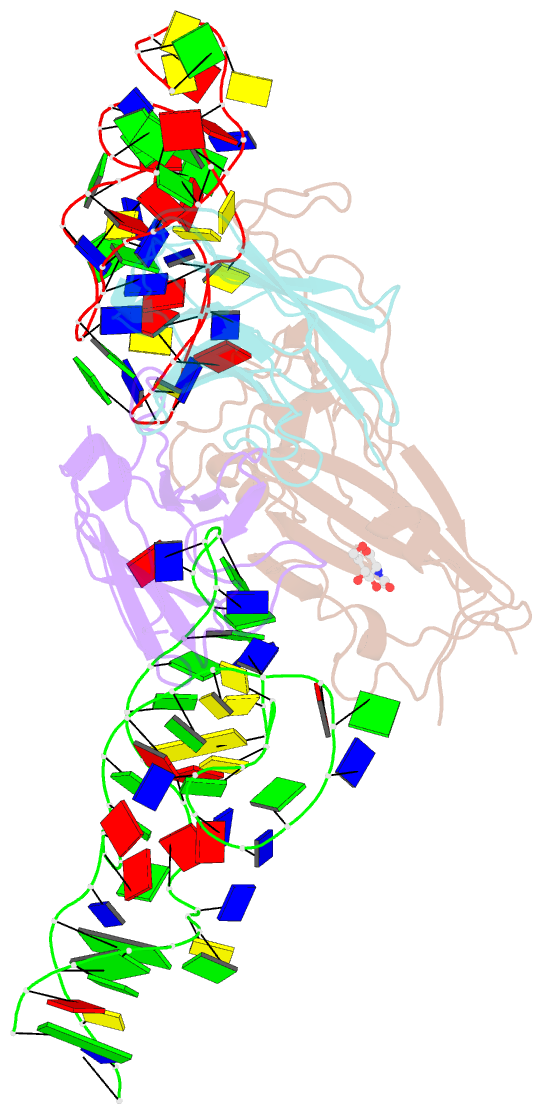
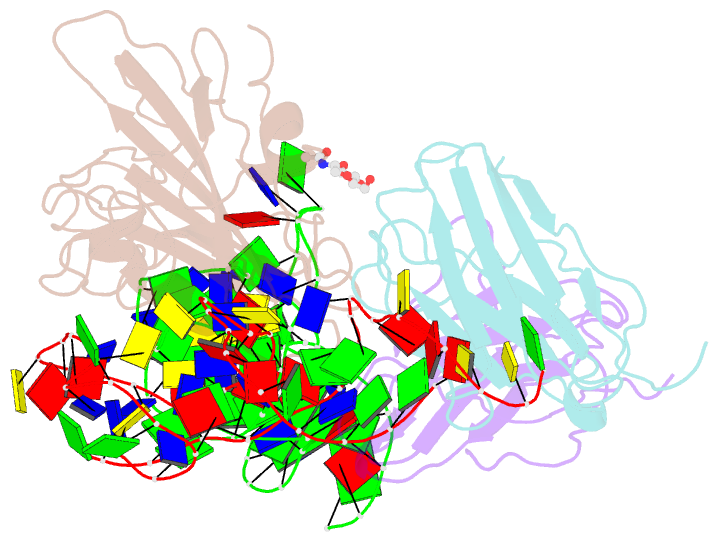
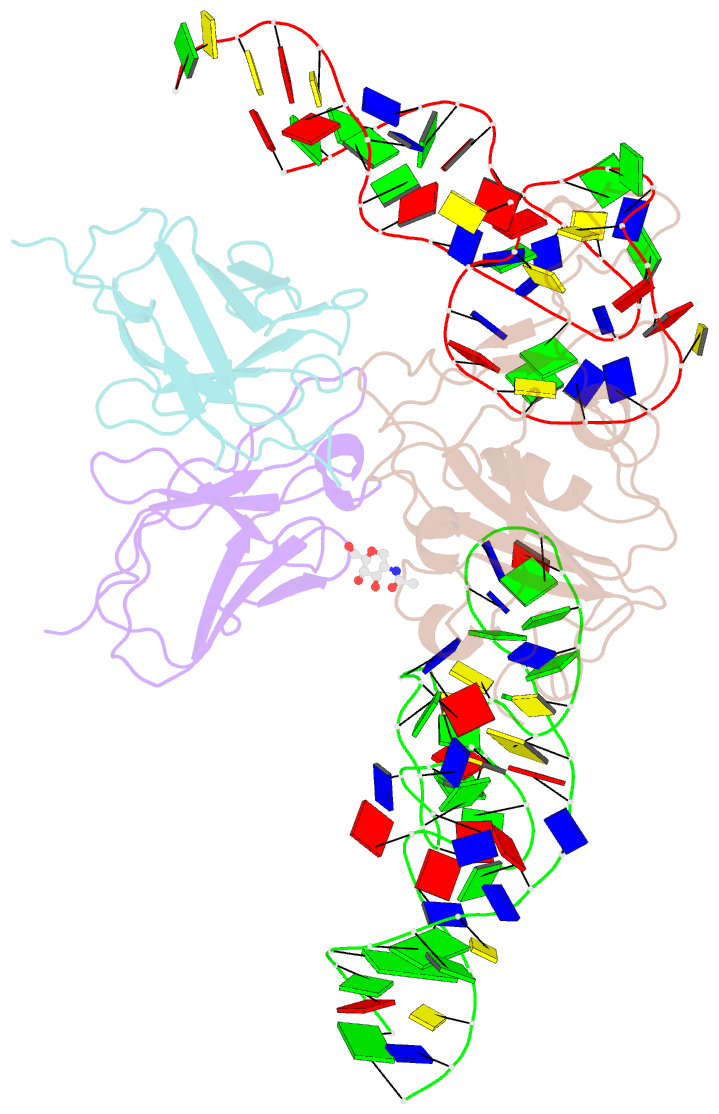
Summary information and primary citation
- PDB-id
- 8j1q; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein-immune system-DNA
- Method
- cryo-EM (3.3 Å)
- Summary
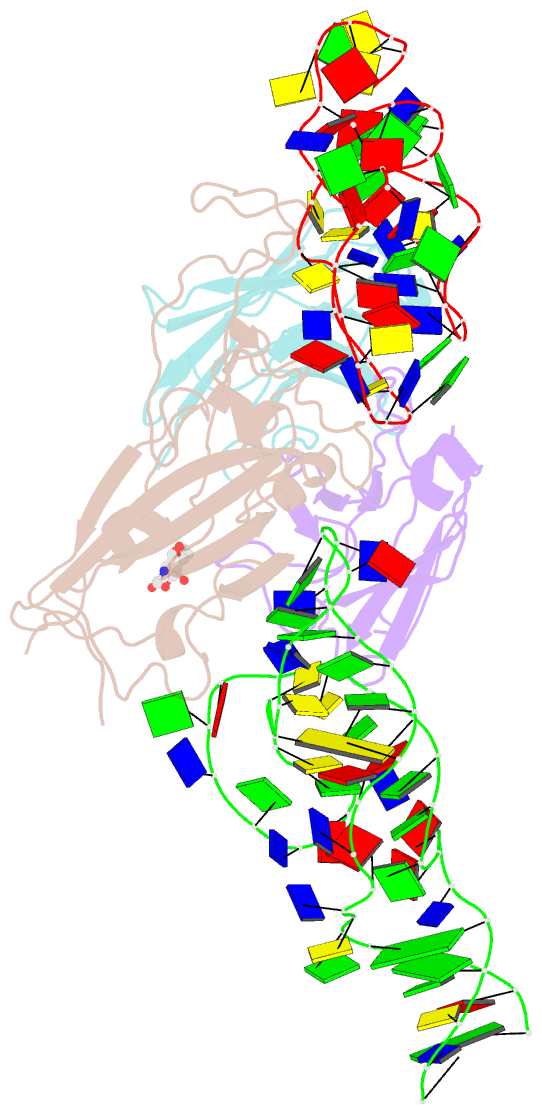
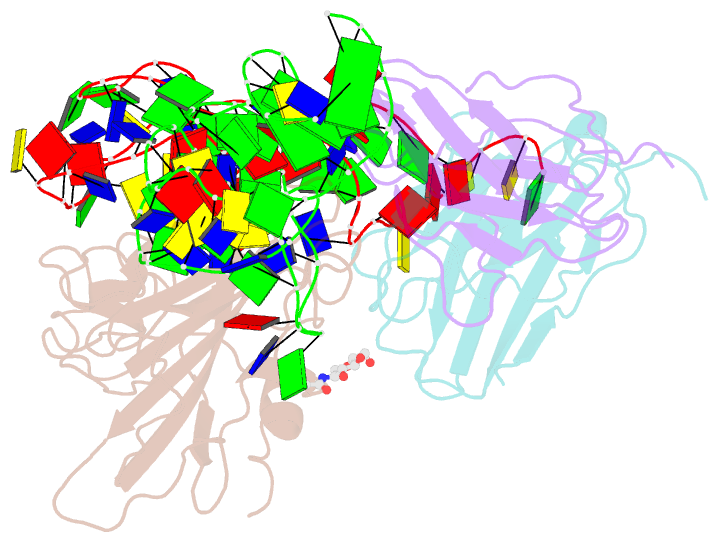
- Cryoem structure of sars cov-2 rbd and aptamer complex
- Reference
- Rahman MS, Han MJ, Kim SW, Kang SM, Kim BR, Kim H, Lee CJ, Noh JE, Kim H, Lee JO, Jang SK (2023): "Structure-Guided Development of Bivalent Aptamers Blocking SARS-CoV-2 Infection." Molecules, 28. doi: 10.3390/molecules28124645.
- Abstract
- Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused devastation to human society through its high virulence, infectivity, and genomic mutations, which reduced the efficacy of vaccines. Here, we report the development of aptamers that effectively interfere with SARS-CoV-2 infection by targeting its spike protein, which plays a pivotal role in host cell entry of the virus through interaction with the viral receptor angiotensin-converting enzyme 2 (ACE2). To develop highly effective aptamers and to understand their mechanism in inhibiting viral infection, we determined the three-dimensional (3D) structures of aptamer/receptor-binding domain (RBD) complexes using cryogenic electron microscopy (cryo-EM). Moreover, we developed bivalent aptamers targeting two distinct regions of the RBD in the spike protein that directly interact with ACE2. One aptamer interferes with the binding of ACE2 by blocking the ACE2-binding site in RBD, and the other aptamer allosterically inhibits ACE2 by binding to a distinct face of RBD. Using the 3D structures of aptamer-RBD complexes, we minimized and optimized these aptamers. By combining the optimized aptamers, we developed a bivalent aptamer that showed a stronger inhibitory effect on virus infection than the component aptamers. This study confirms that the structure-based aptamer-design approach has a high potential in developing antiviral drugs against SARS-CoV-2 and other viruses.