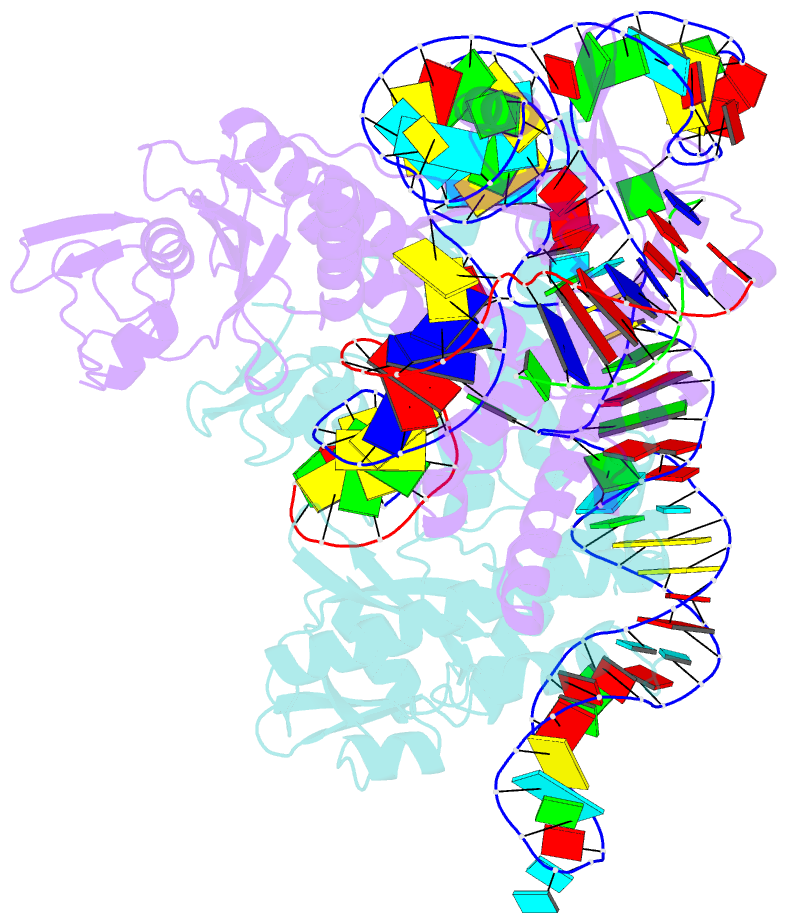
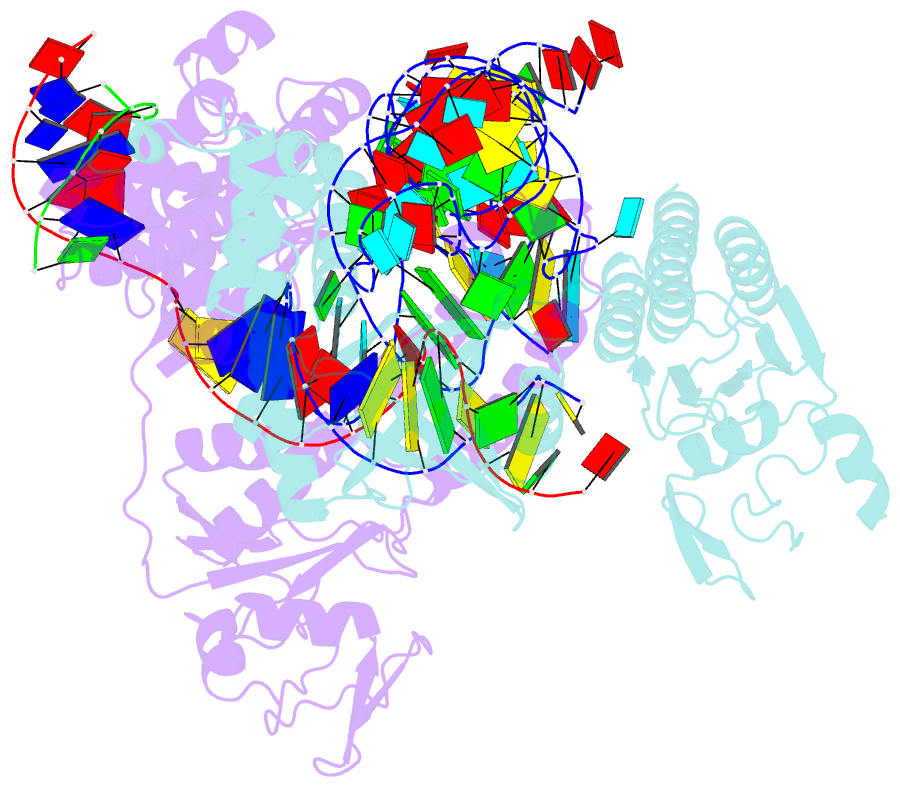
Summary information and primary citation
- PDB-id
- 8j3r; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-DNA-RNA
- Method
- cryo-EM (2.95 Å)
- Summary
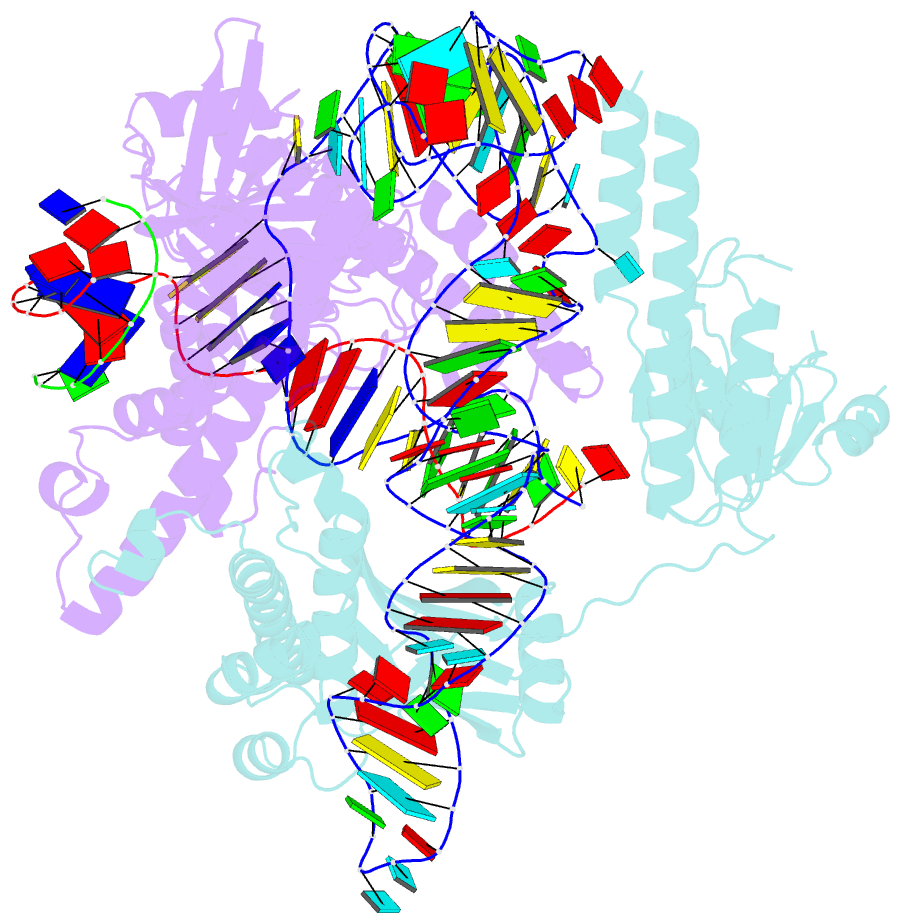
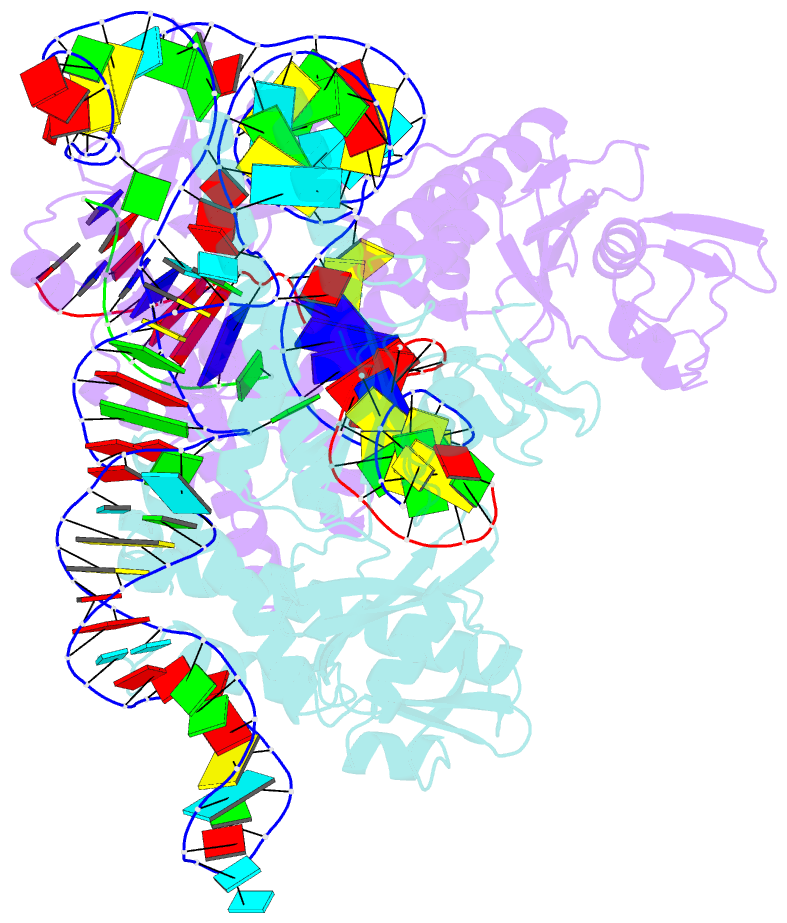
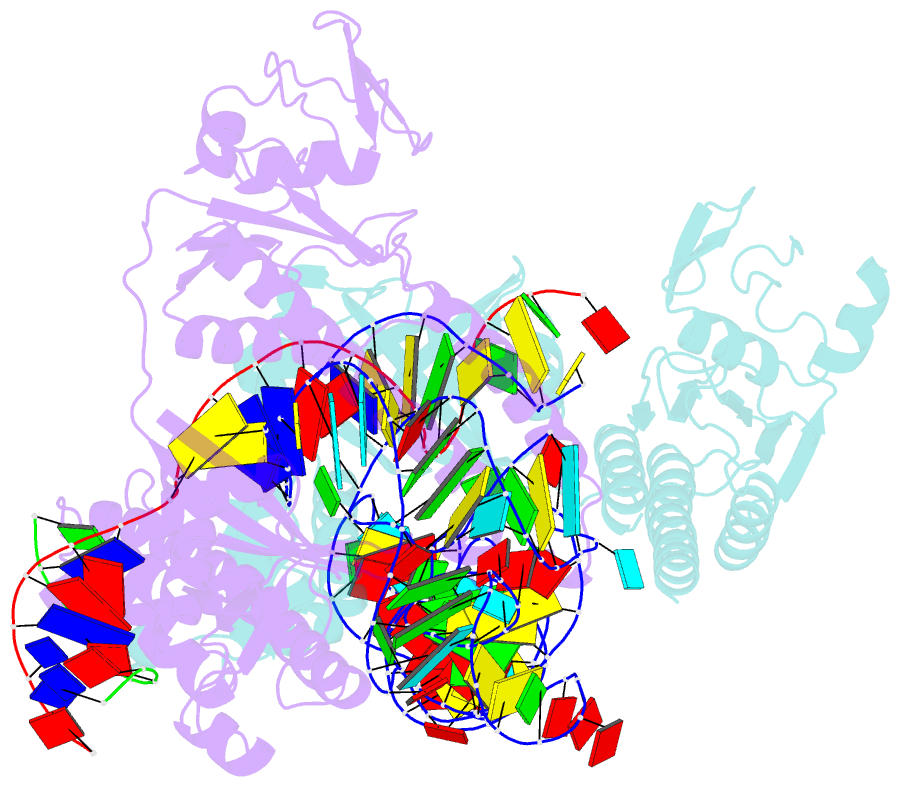
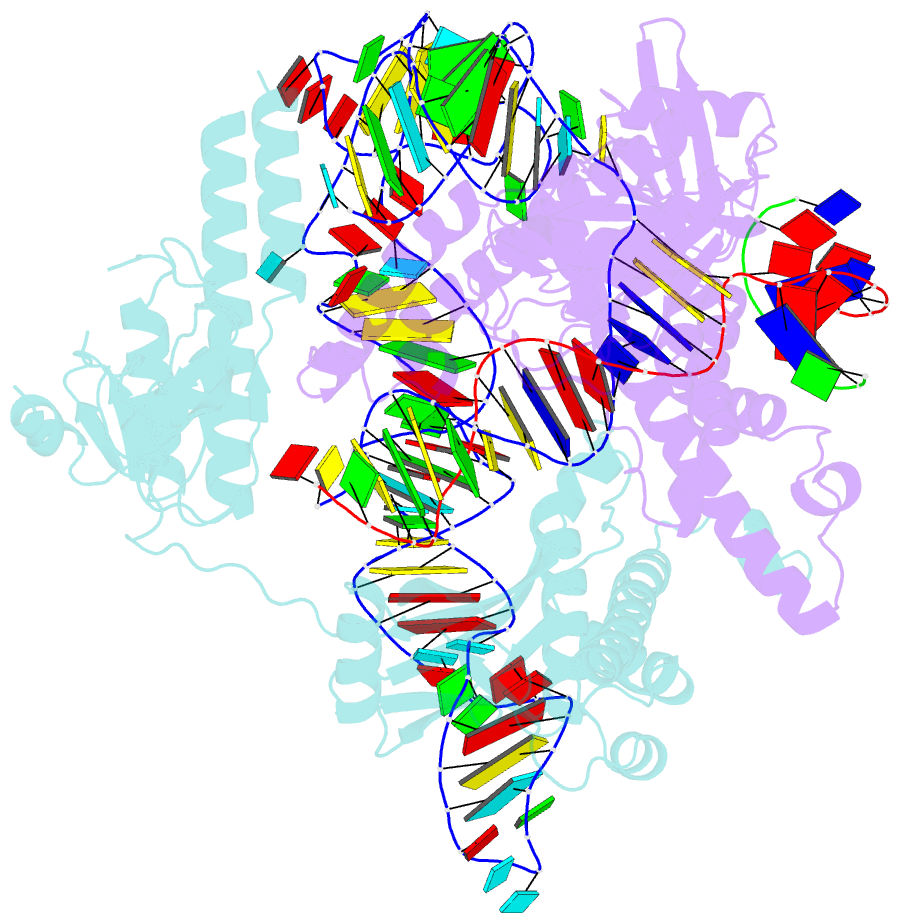
- cryo-EM structure of the ascas12f-hkra-sgrnas3-5v7-target DNA
- Reference
- Hino T, Omura SN, Nakagawa R, Togashi T, Takeda SN, Hiramoto T, Tasaka S, Hirano H, Tokuyama T, Uosaki H, Ishiguro S, Kagieva M, Yamano H, Ozaki Y, Motooka D, Mori H, Kirita Y, Kise Y, Itoh Y, Matoba S, Aburatani H, Yachie N, Karvelis T, Siksnys V, Ohmori T, Hoshino A, Nureki O (2023): "An AsCas12f-based compact genome-editing tool derived by deep mutational scanning and structural analysis." Cell, 186, 4920-4935.e23. doi: 10.1016/j.cell.2023.08.031.
- Abstract
- SpCas9 and AsCas12a are widely utilized as genome-editing tools in human cells. However, their relatively large size poses a limitation for delivery by cargo-size-limited adeno-associated virus (AAV) vectors. The type V-F Cas12f from Acidibacillus sulfuroxidans is exceptionally compact (422 amino acids) and has been harnessed as a compact genome-editing tool. Here, we developed an approach, combining deep mutational scanning and structure-informed design, to successfully generate two AsCas12f activity-enhanced (enAsCas12f) variants. Remarkably, the enAsCas12f variants exhibited genome-editing activities in human cells comparable with those of SpCas9 and AsCas12a. The cryoelectron microscopy (cryo-EM) structures revealed that the mutations stabilize the dimer formation and reinforce interactions with nucleic acids to enhance their DNA cleavage activities. Moreover, enAsCas12f packaged with partner genes in an all-in-one AAV vector exhibited efficient knock-in/knock-out activities and transcriptional activation in mice. Taken together, enAsCas12f variants could offer a minimal genome-editing platform for in vivo gene therapy.