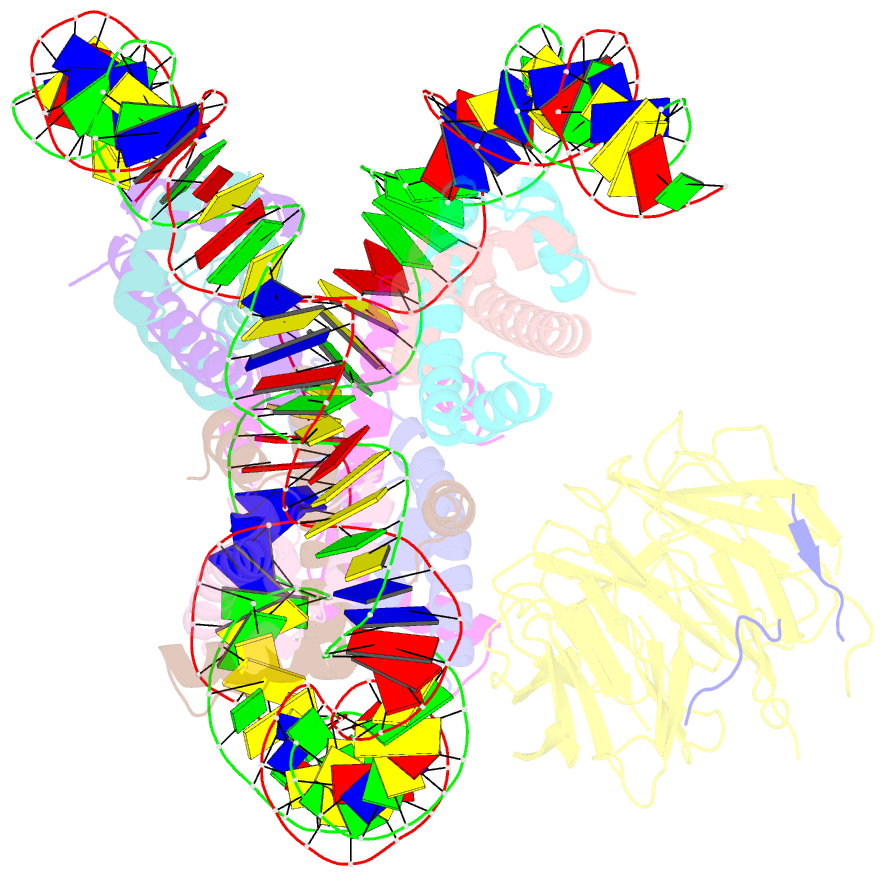
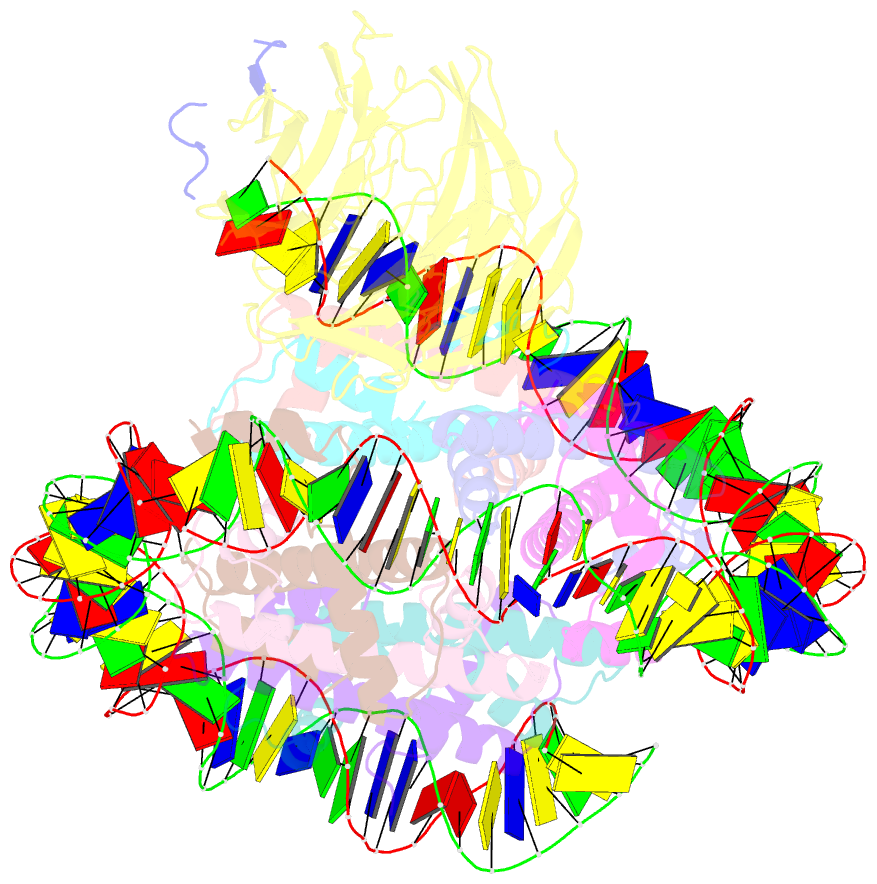
Summary information and primary citation
- PDB-id
- 8j6s; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- replication-DNA
- Method
- cryo-EM (3.8 Å)
- Summary
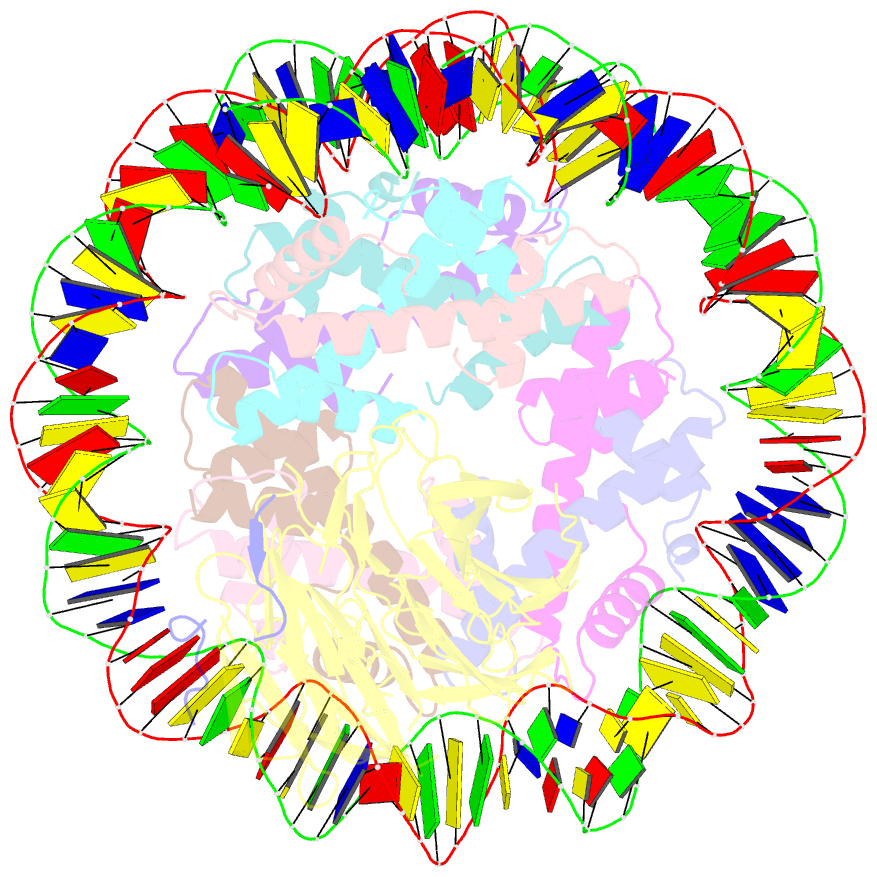
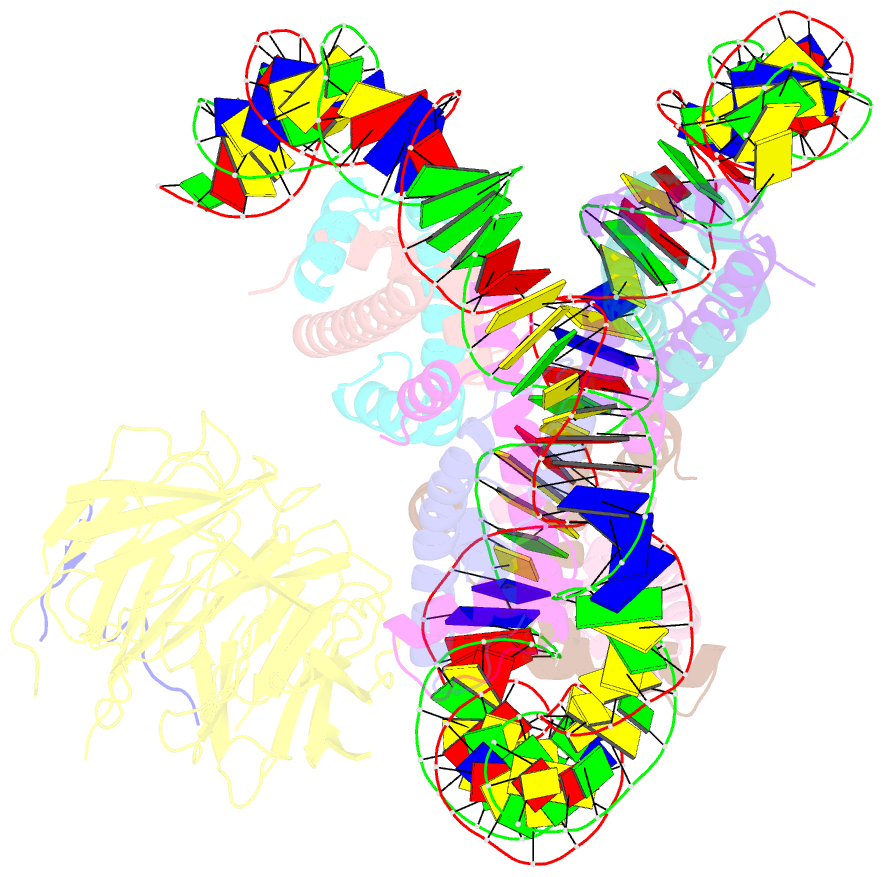
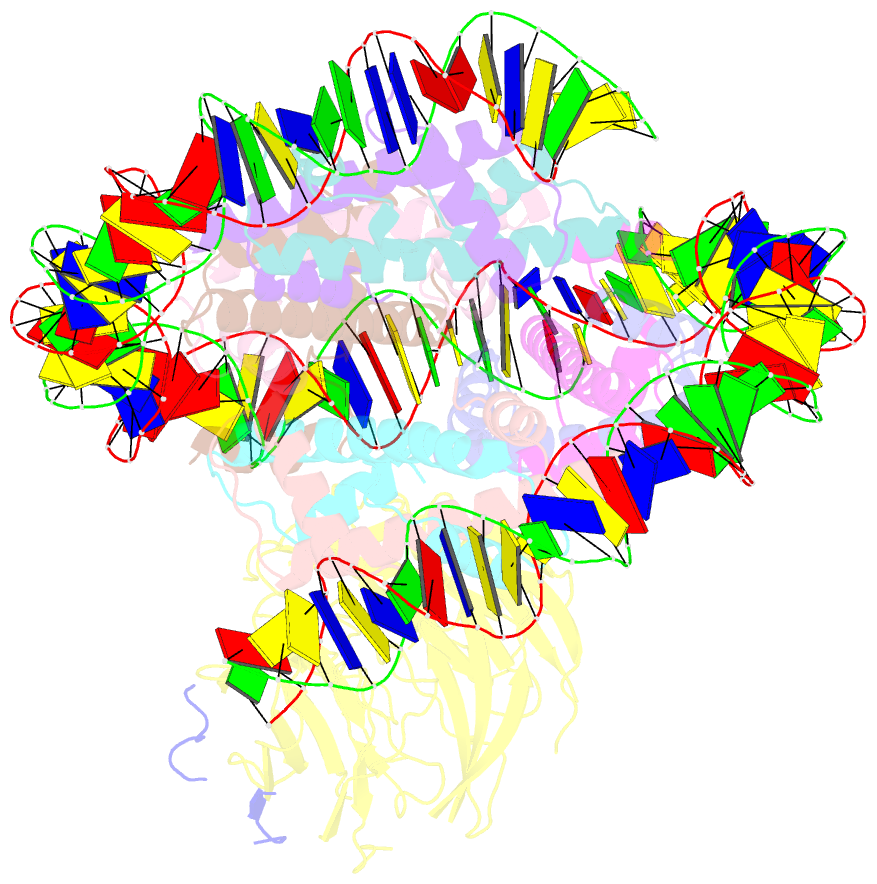
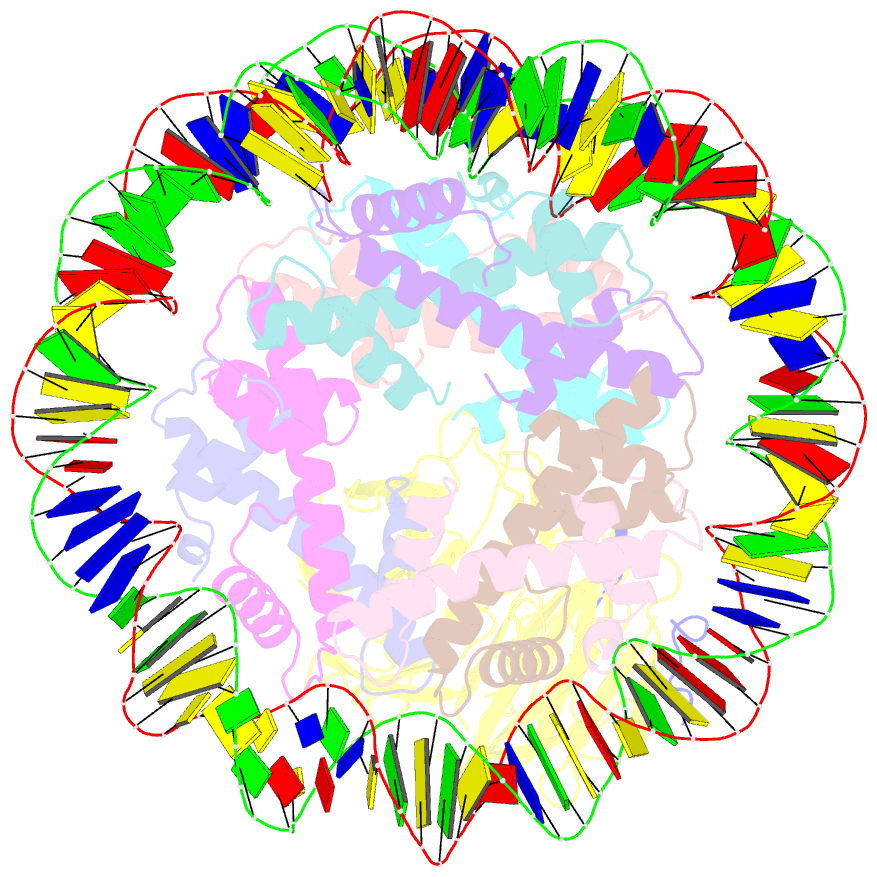
- cryo-EM structure of the single caf-1 bound right-handed di-tetrasome
- Reference
- Liu CP, Yu Z, Xiong J, Hu J, Song A, Ding D, Yu C, Yang N, Wang M, Yu J, Hou P, Zeng K, Li Z, Zhang Z, Zhang X, Li W, Zhang Z, Zhu B, Li G, Xu RM (2023): "Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1." Science, 381, eadd8673. doi: 10.1126/science.add8673.
- Abstract
- Chromatin inheritance entails de novo nucleosome assembly after DNA replication by chromatin assembly factor-1 (CAF-1). Yet direct knowledge about CAF-1's histone binding mode and nucleosome assembly process is lacking. In this work, we report the crystal structure of human CAF-1 in the absence of histones and the cryo-electron microscopy structure of CAF-1 in complex with histones H3 and H4. One histone H3-H4 heterodimer is bound by one CAF-1 complex mainly through the p60 subunit and the acidic domain of the p150 subunit. We also observed a dimeric CAF-1-H3-H4 supercomplex in which two H3-H4 heterodimers are poised for tetramer assembly and discovered that CAF-1 facilitates right-handed DNA wrapping of H3-H4 tetramers. These findings signify the involvement of DNA in H3-H4 tetramer formation and suggest a right-handed nucleosome precursor in chromatin replication.