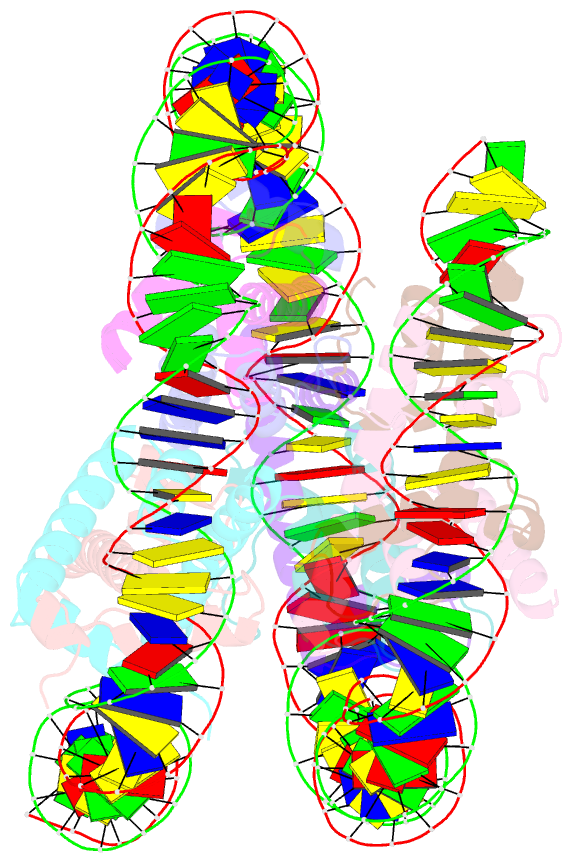
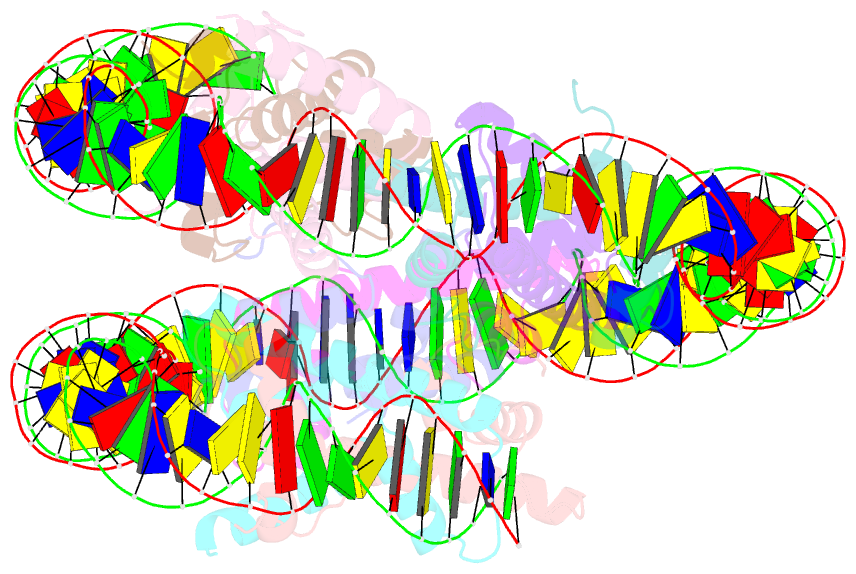
Summary information and primary citation
- PDB-id
- 8j91; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- nuclear protein
- Method
- cryo-EM (2.9 Å)
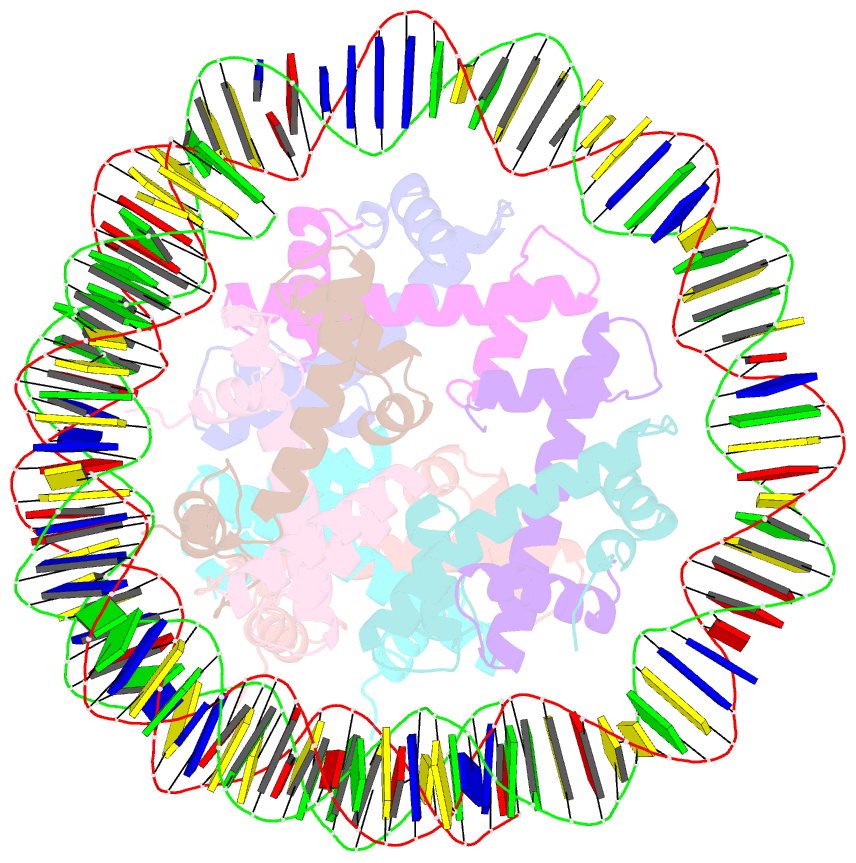
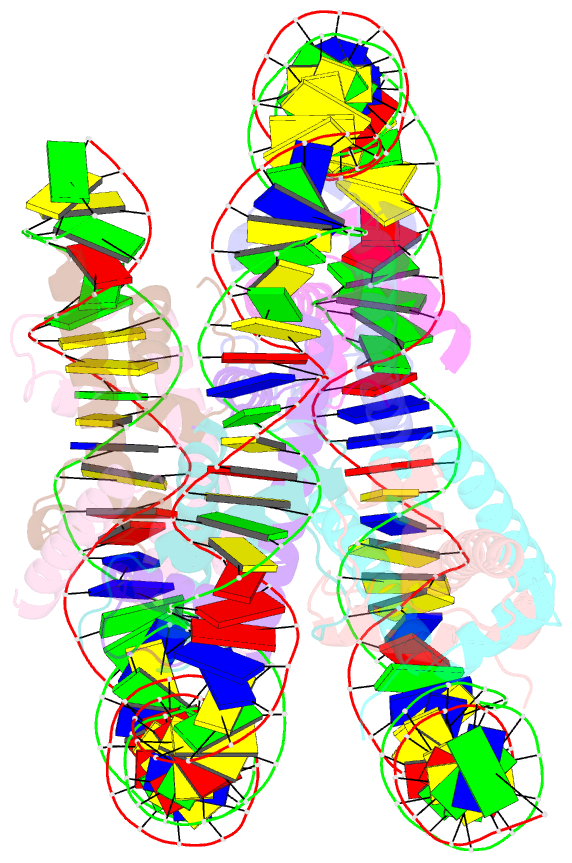
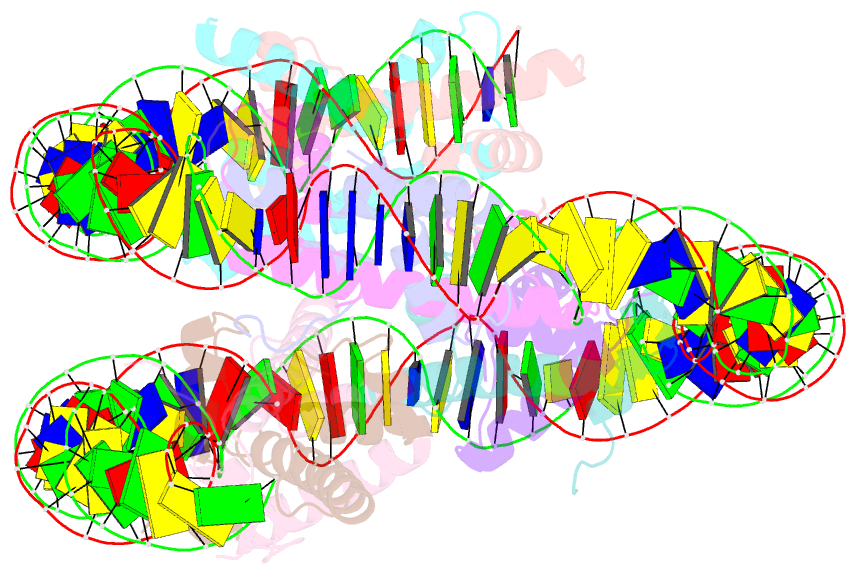
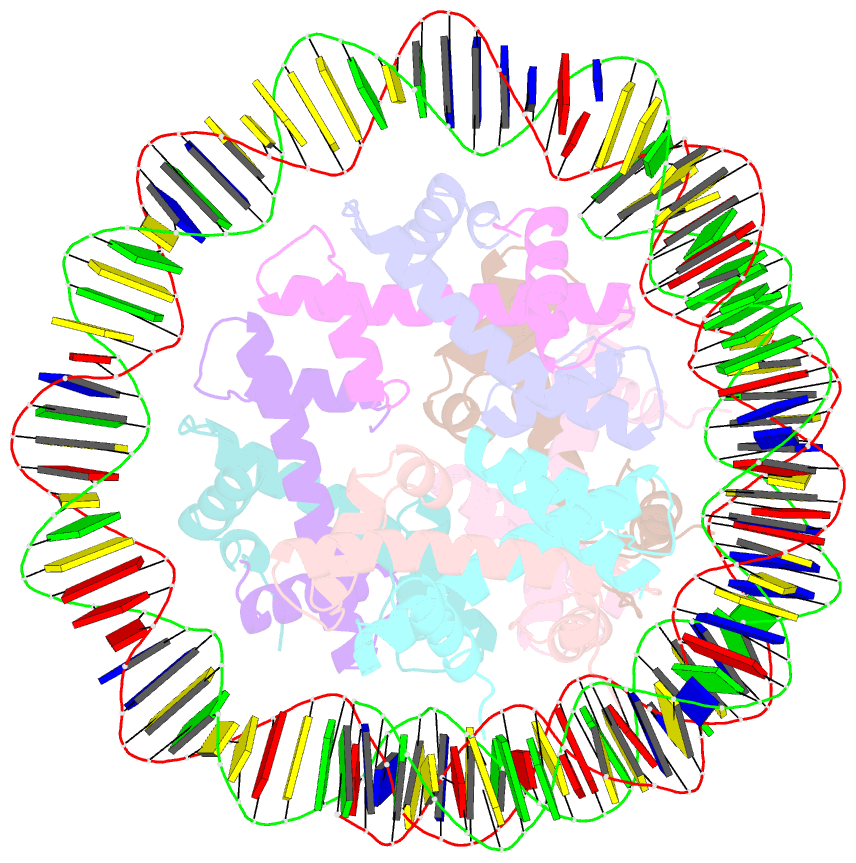
- Summary
- cryo-EM structure of nucleosome containing arabidopsis thaliana histones
- Reference
- Osakabe A, Takizawa Y, Horikoshi N, Hatazawa S, Negishi L, Sato S, Berger F, Kakutani T, Kurumizaka H (2024): "Molecular and structural basis of the chromatin remodeling activity by Arabidopsis DDM1." Nat Commun, 15, 5187. doi: 10.1038/s41467-024-49465-w.
- Abstract
- The histone H2A variant H2A.W occupies transposons and thus prevents access to them in Arabidopsis thaliana. H2A.W is deposited by the chromatin remodeler DDM1, which also promotes the accessibility of chromatin writers to heterochromatin by an unknown mechanism. To shed light on this question, we solve the cryo-EM structures of nucleosomes containing H2A and H2A.W, and the DDM1-H2A.W nucleosome complex. These structures show that the DNA end flexibility of the H2A nucleosome is higher than that of the H2A.W nucleosome. In the DDM1-H2A.W nucleosome complex, DDM1 binds to the N-terminal tail of H4 and the nucleosomal DNA and increases the DNA end flexibility of H2A.W nucleosomes. Based on these biochemical and structural results, we propose that DDM1 counters the low accessibility caused by nucleosomes containing H2A.W to enable the maintenance of repressive epigenetic marks on transposons and prevent their activity.