Summary information and primary citation
- PDB-id
- 8jmk; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- X-ray (2.7 Å)
- Summary
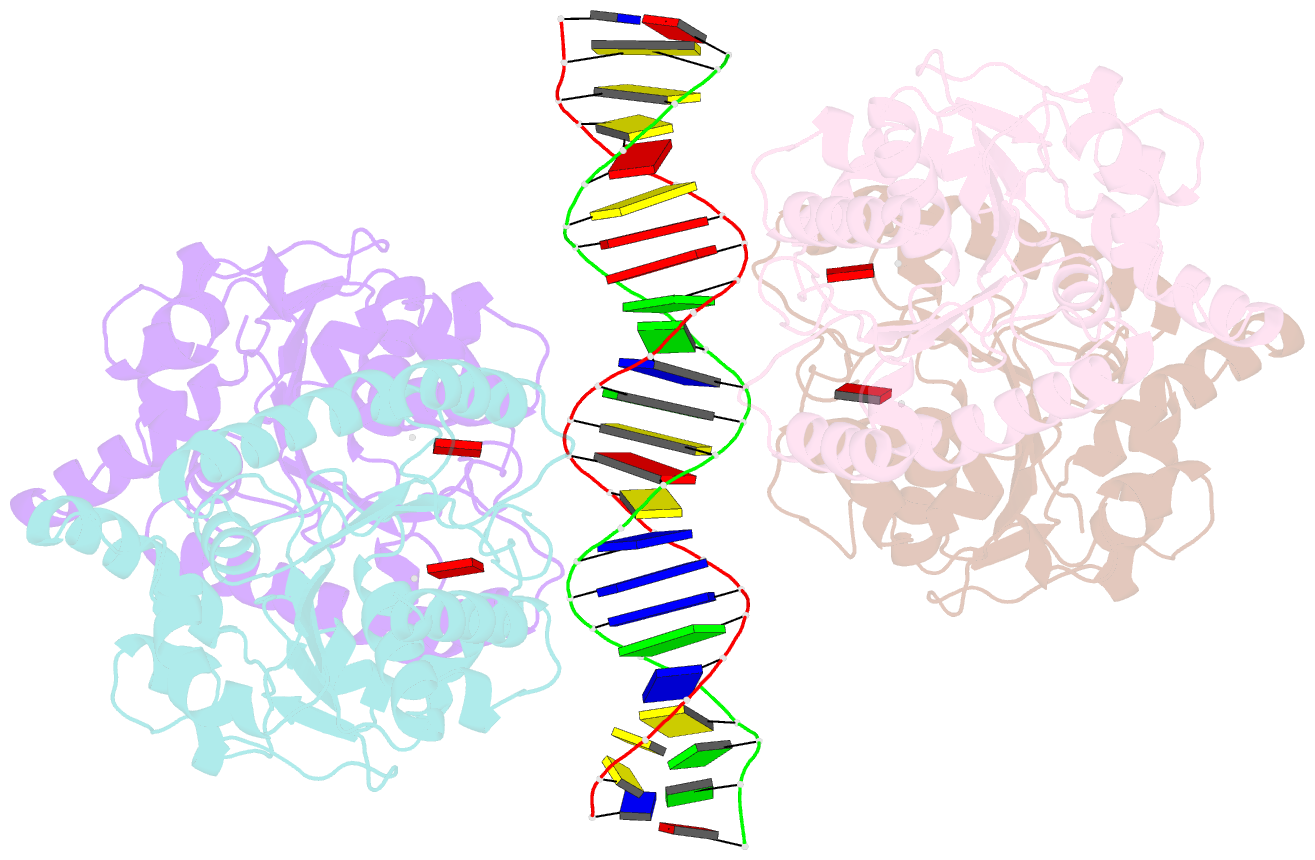
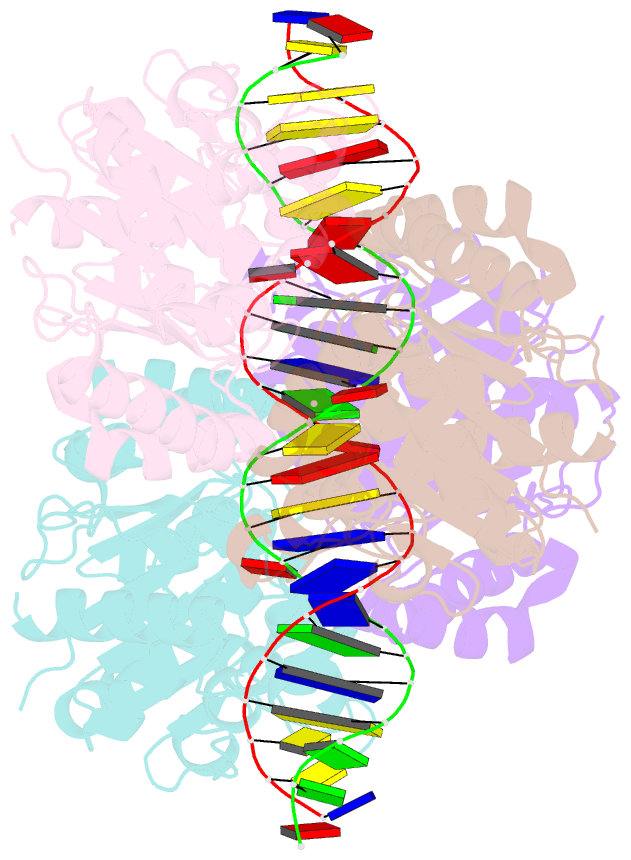
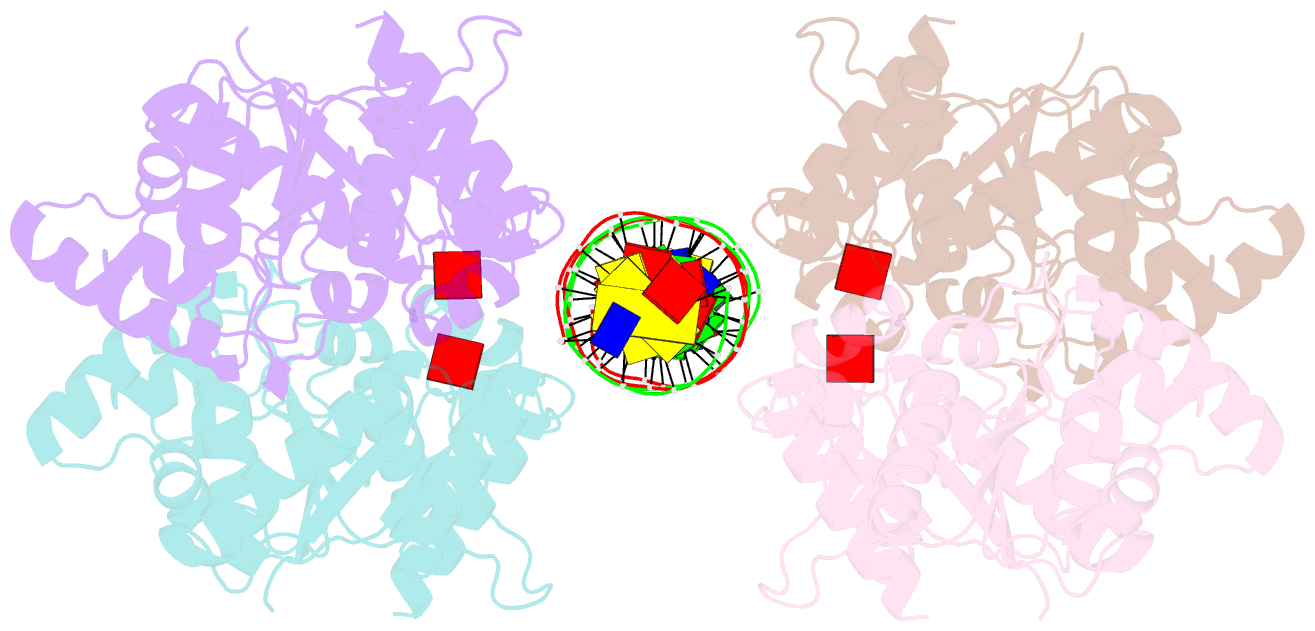
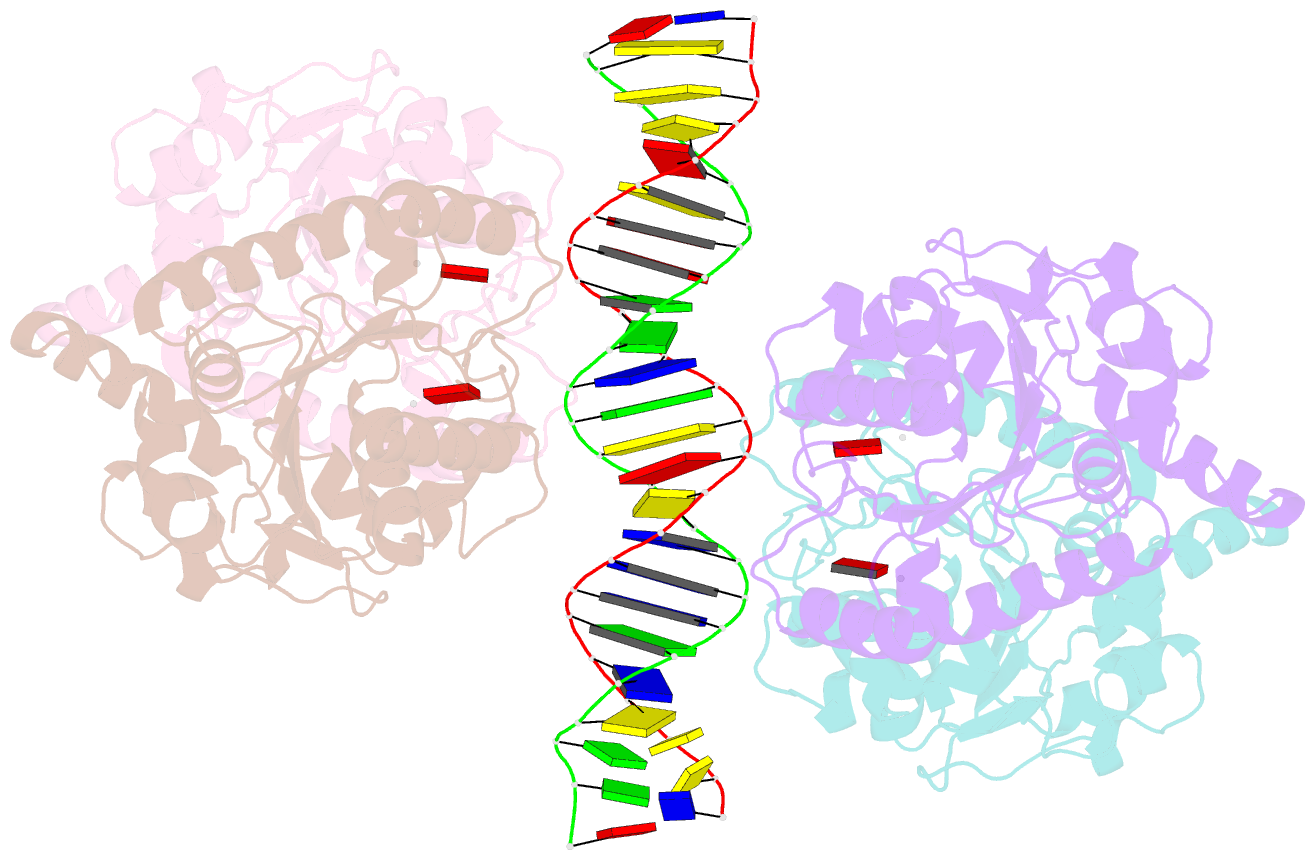
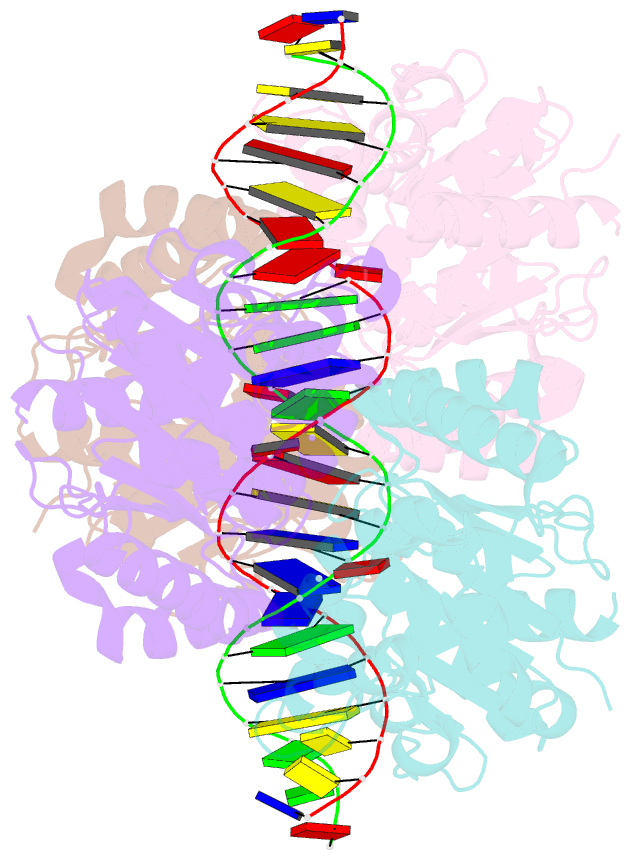
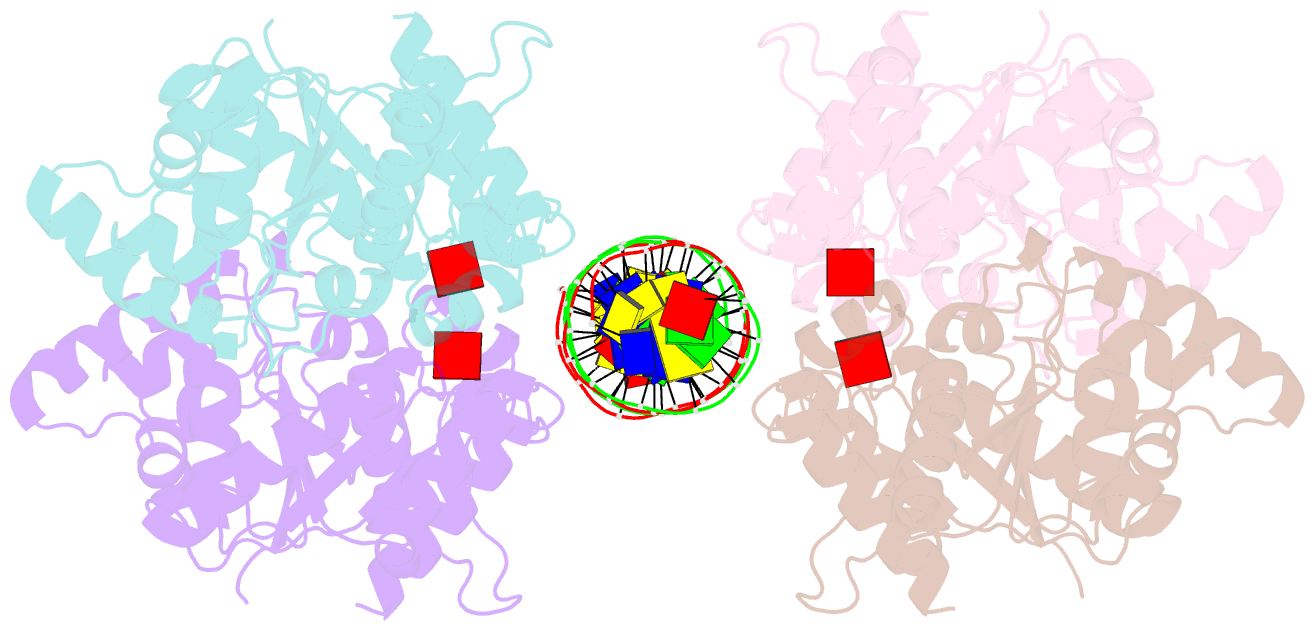
- Structure of helicobacter pylori soj mutant, d41a bound to DNA
- Reference
- Chu CH, Wu CT, Lin MG, Yen CY, Wu YZ, Hsiao CD, Sun YJ (2024): "Insights into the molecular mechanism of ParABS system in chromosome partition by HpParA and HpParB." Nucleic Acids Res., 52, 7321-7336. doi: 10.1093/nar/gkae450.
- Abstract
- The ParABS system, composed of ParA (an ATPase), ParB (a DNA binding protein), and parS (a centromere-like DNA), regulates bacterial chromosome partition. The ParB-parS partition complex interacts with the nucleoid-bound ParA to form the nucleoid-adaptor complex (NAC). In Helicobacter pylori, ParA and ParB homologs are encoded as HpSoj and HpSpo0J (HpParA and HpParB), respectively. We determined the crystal structures of the ATP hydrolysis deficient mutant, HpParAD41A, and the HpParAD41A-DNA complex. We assayed the CTPase activity of HpParB and identified two potential DNA binding modes of HpParB regulated by CTP, one is the specific DNA binding by the DNA binding domain and the other is the non-specific DNA binding through the C-terminal domain under the regulation of CTP. We observed an interaction between HpParAD41A and the N-terminus fragment of HpParB (residue 1-10, HpParBN10) and determined the crystal structure of the ternary complex, HpParAD41A-DNA-HpParBN10 complex which mimics the NAC formation. HpParBN10 binds near the HpParAD41A dimer interface and is clamped by flexible loops, L23 and L34, through a specific cation-π interaction between Arg9 of HpParBN10 and Phe52 of HpParAD41A. We propose a molecular mechanism model of the ParABS system providing insight into chromosome partition in bacteria.