Summary information and primary citation
- PDB-id
- 8joz; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-RNA
- Method
- X-ray (2.22 Å)
- Summary
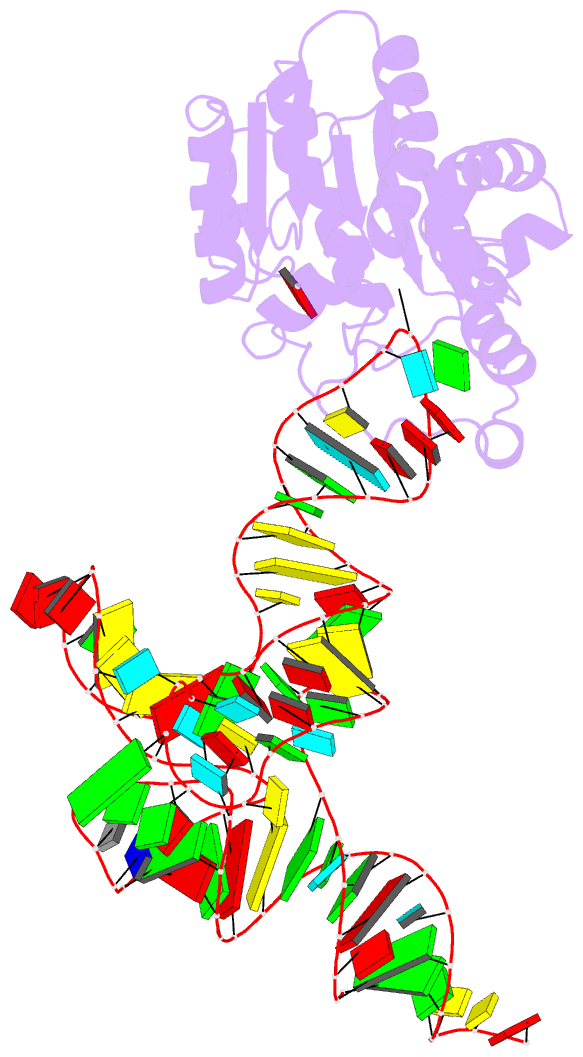
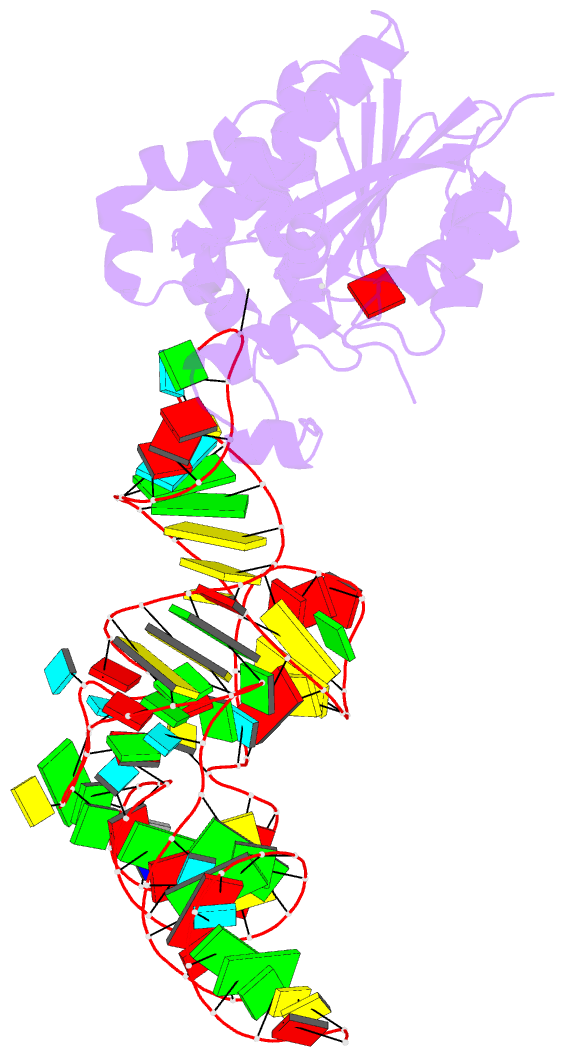
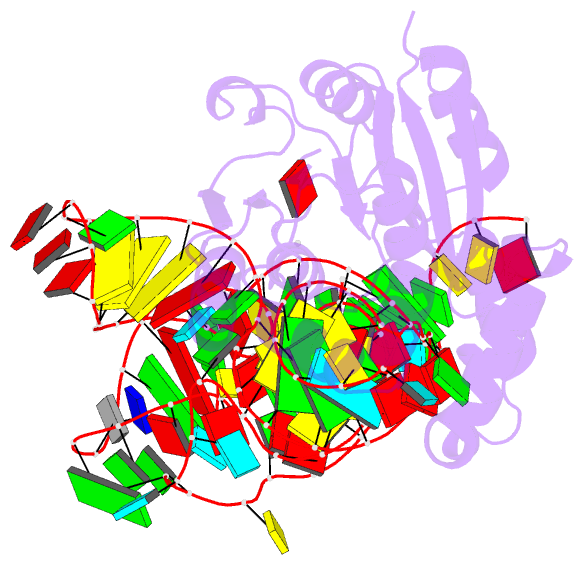
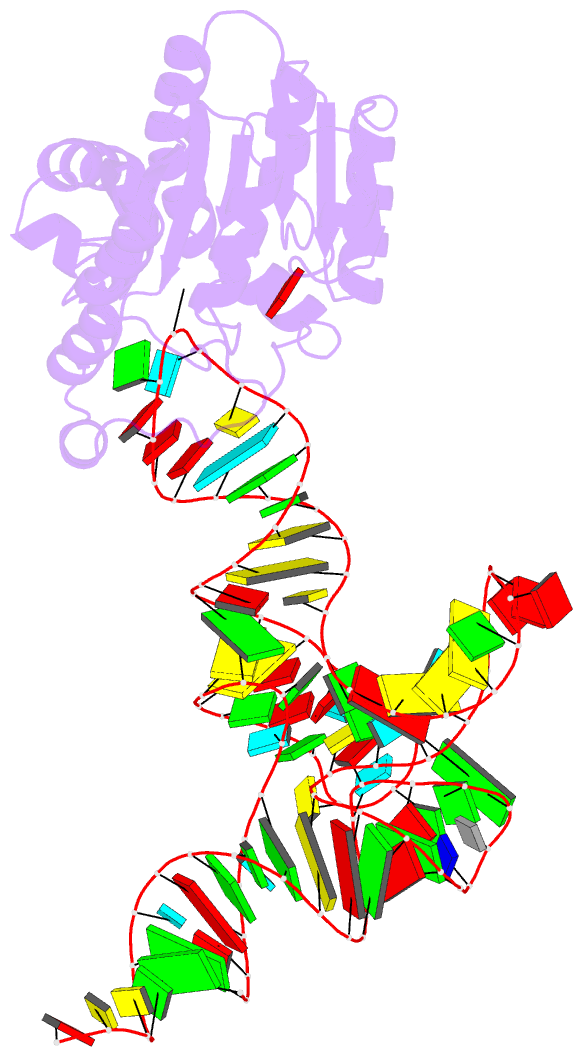
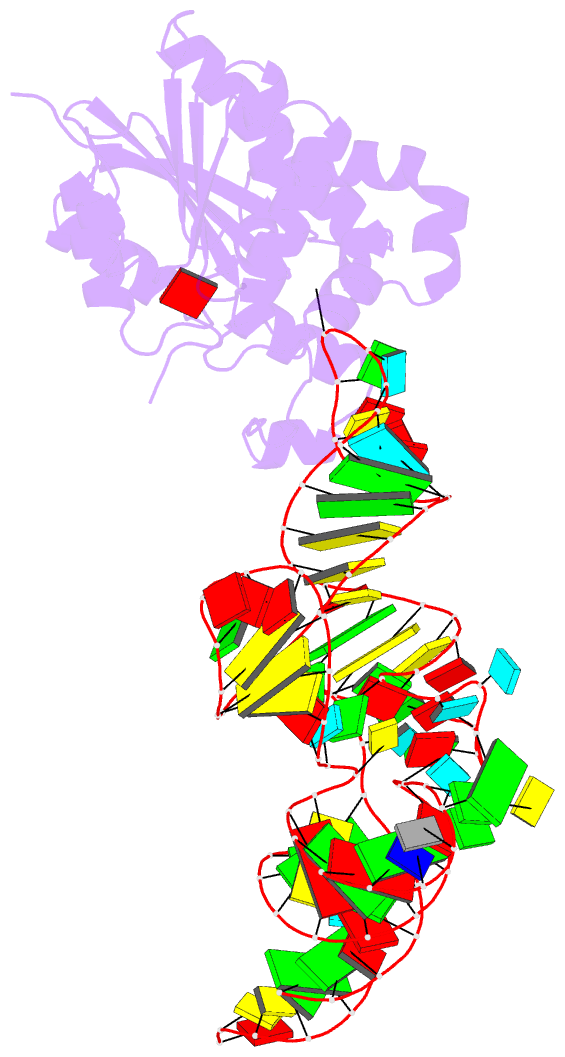
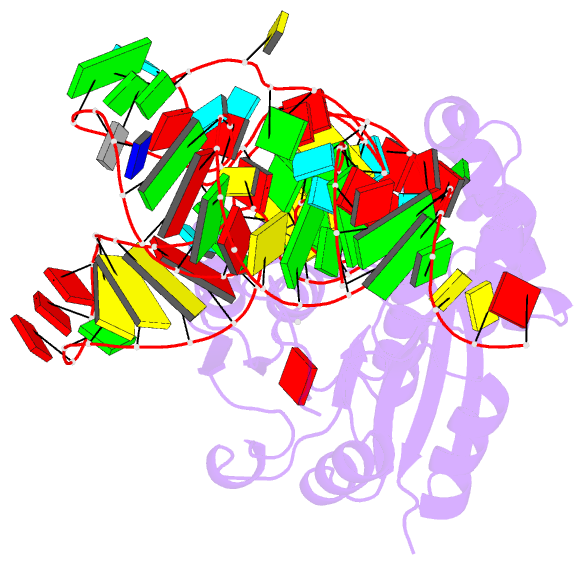
- Crystal structure of cmom from e. coli complexed with sinefungin and cellularly expressed trna ser
- Reference
- Yoo J, Lee J, Kim J (2023): "Structural basis for the selective methylation of 5-carboxymethoxyuridine in tRNA modification." Nucleic Acids Res., 51, 9432-9441. doi: 10.1093/nar/gkad668.
- Abstract
- Posttranscriptional modifications of tRNA are widely conserved in all domains of life. Especially, those occurring within the anticodon often modulate translational efficiency. Derivatives of 5-hydroxyuridine are specifically found in bacterial tRNA, where 5-methoxyuridine and 5-carboxymethoxyuridine are the major species in Gram-positive and Gram-negative bacteria, respectively. In certain tRNA species, 5-carboxymethoxyuridine can be further methylated by CmoM to form the methyl ester. In this report, we present the X-ray crystal structure of Escherichia coli CmoM complexed with tRNASer1, which contains 5-carboxymethoxyuridine at the 5'-end of anticodon (the 34th position of tRNA). The 2.22 Å resolution structure of the enzyme-tRNA complex reveals that both the protein and tRNA undergo local conformational changes around the binding interface. Especially, the hypomodified uracil base is flipped out from the canonical stacked conformation enabling the specific molecular interactions with the enzyme. Moreover, the structure illustrates that the enzyme senses exclusively the anticodon arm region of the substrate tRNA and examines the presence of key determinants, 5-carboxymethoxyuridine at position 34 and guanosine at position 35, offering molecular basis for the discriminatory mechanism against non-cognate tRNAs.