Summary information and primary citation
- PDB-id
- 8jsg; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- translation
- Method
- cryo-EM (4.6 Å)
- Summary
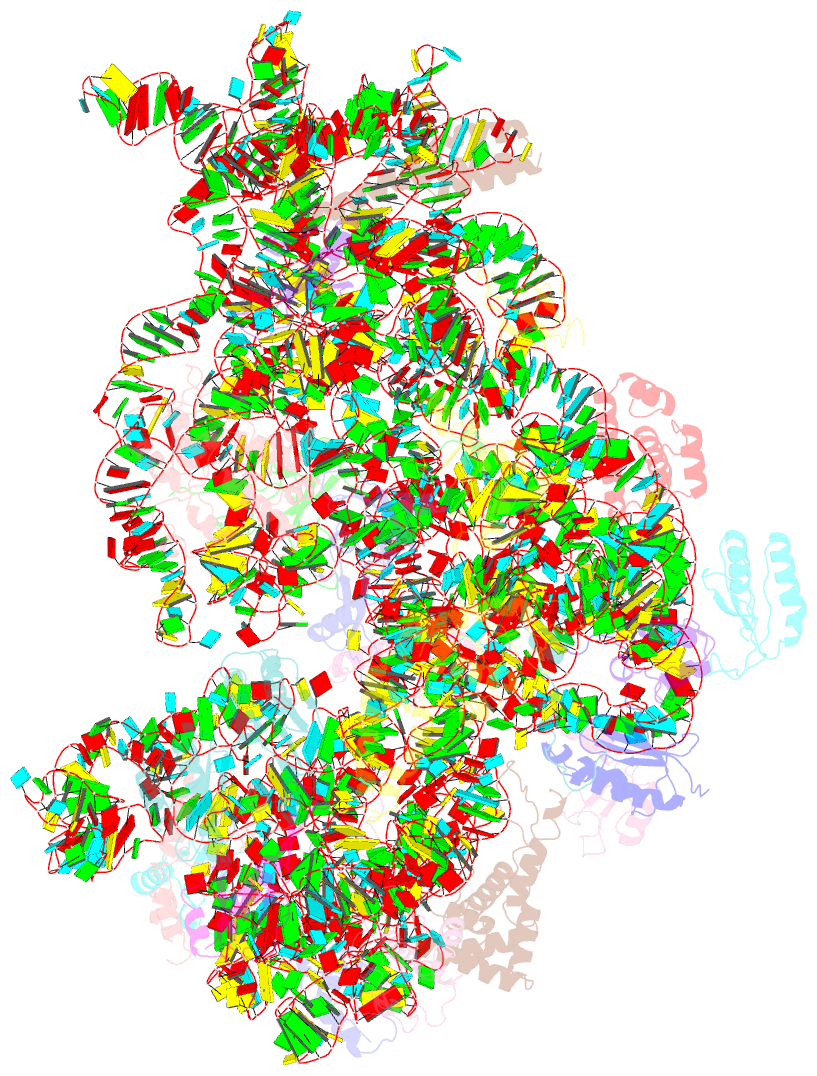
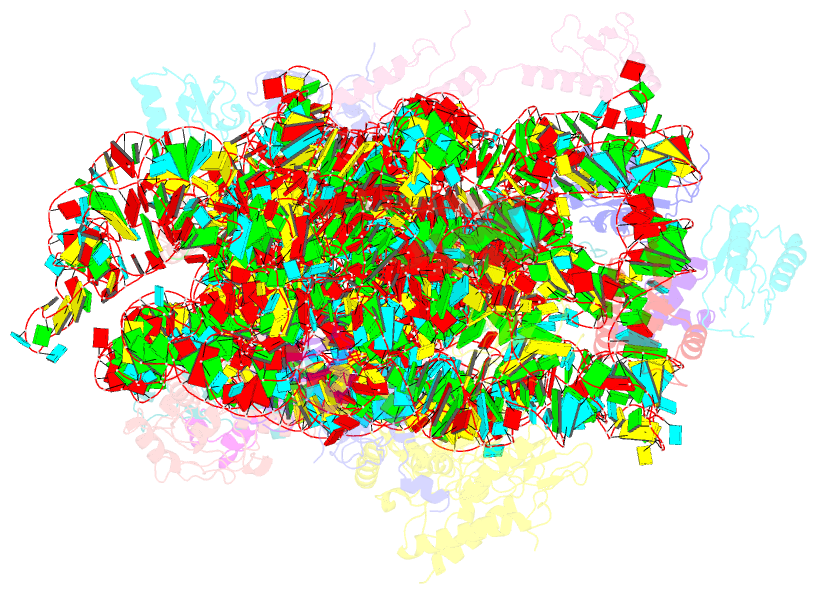
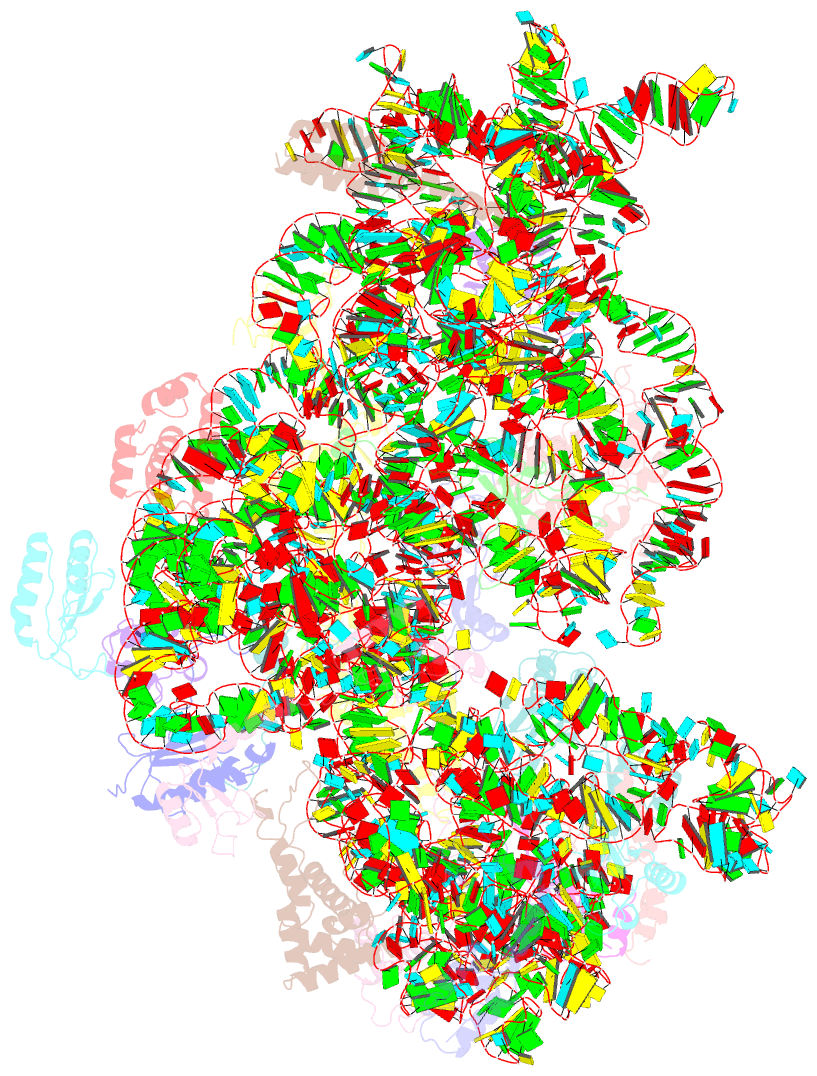
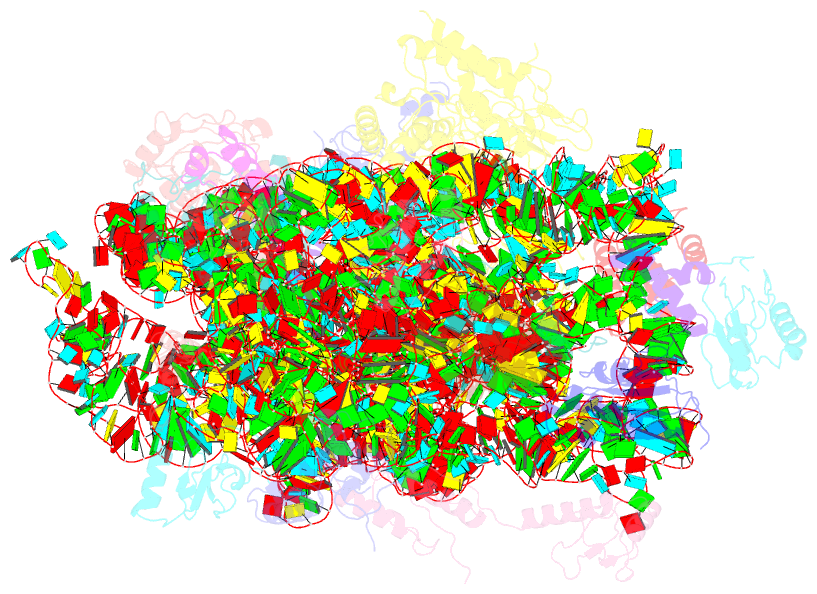
- Structure of the 30s-if3 complex from escherichia coli
- Reference
- Uday AB, Mishra RK, Hussain T (2023): "Initiation factor 3 bound to the 30S ribosomal subunit in an initial step of translation." Proteins. doi: 10.1002/prot.26655.
- Abstract
- Bacterial ribosomes require three initiation factors IF1, IF2, and IF3 during the initial steps of translation. These IFs ensure correct base pairing of the initiator tRNA anticodon with the start codon in the mRNA located at the P-site of the 30S ribosomal subunit. IF3 is one of the first IFs to bind to the 30S and plays a crucial role in the selection of the correct start codon and codon: anticodon base pairing. IF3 also prevents the premature association of the 50S subunit of ribosomes and aids in ribosome recycling. IF3 is reported to change binding sites and conformation to ensure translation initiation fidelity. A recent study suggested an initial binding of IF3 CTD away from the P-site and that IF1 and IF2 promote the movement of CTD to the P-site and concomitant movement of NTD. Hence, to visualize the position of IF3 in the absence of any other IFs, we determined cryo-EM structure of the 30S-IF3 complex. The map shows that IF3 is present in an extended conformation with CTD present at the P-site and NTD near the platform even in the absence of IF1 and IF2. Hence, IF3 CTD binds at the P-site and moves away during the accommodation of the initiator tRNA at the P-site in the later steps of translation initiation. Overall, we report the structure of 30S-IF3 which demystifies the starting binding site and conformation of IF3 on the 30S ribosomal subunit.