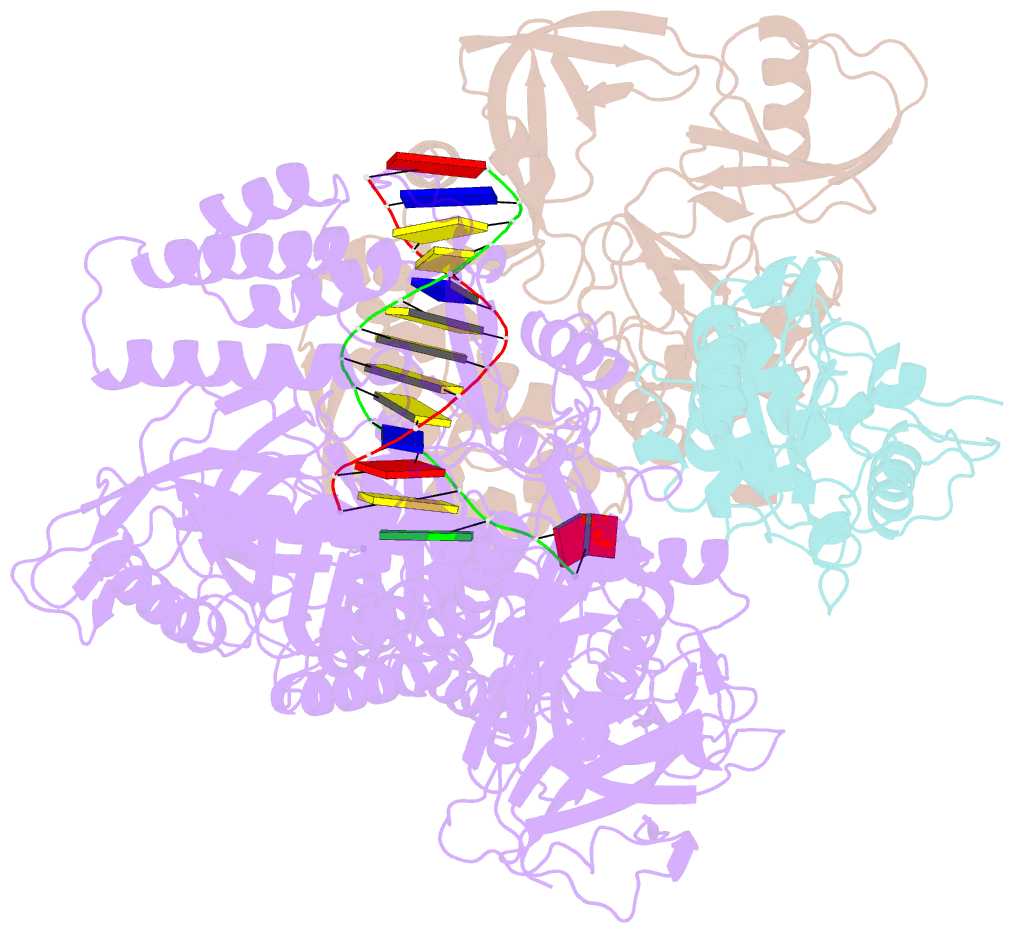
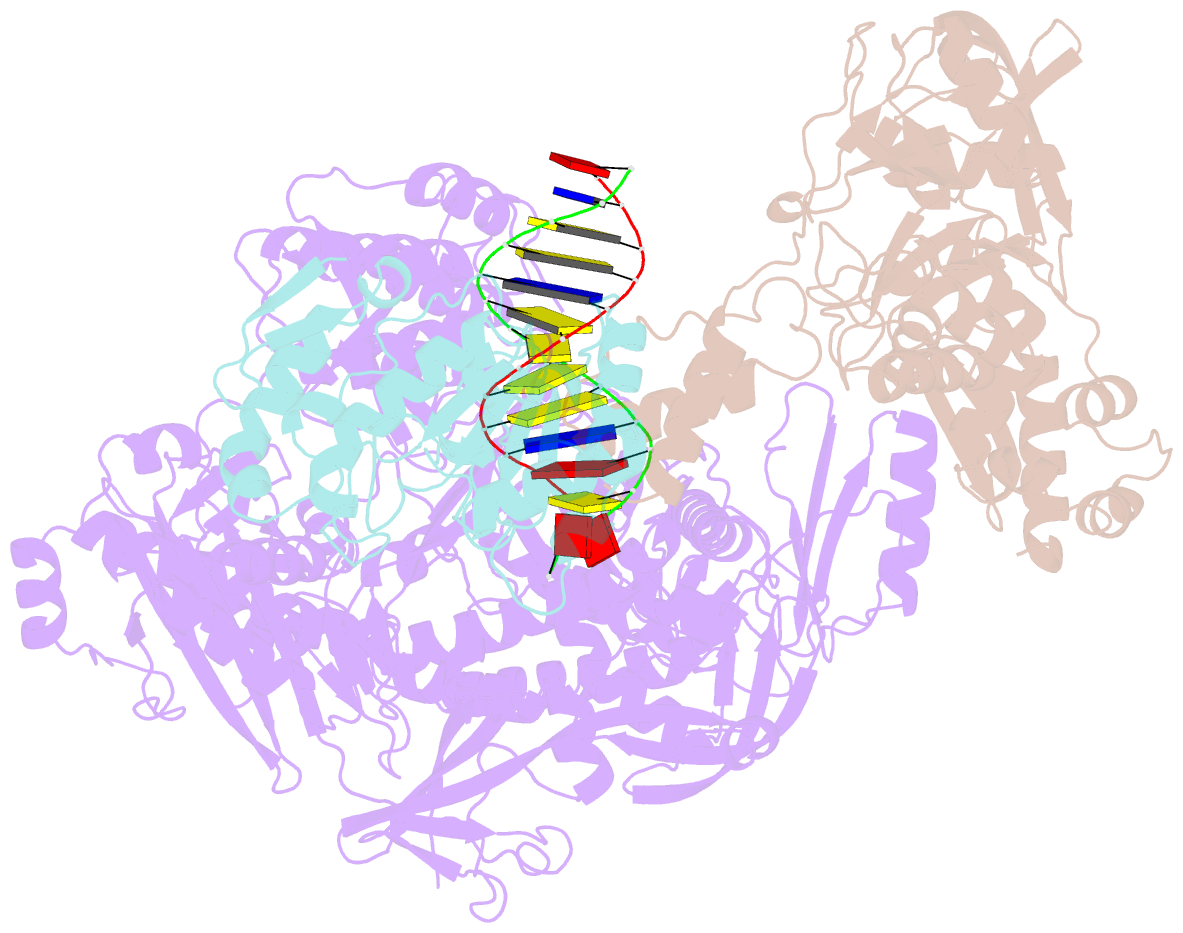
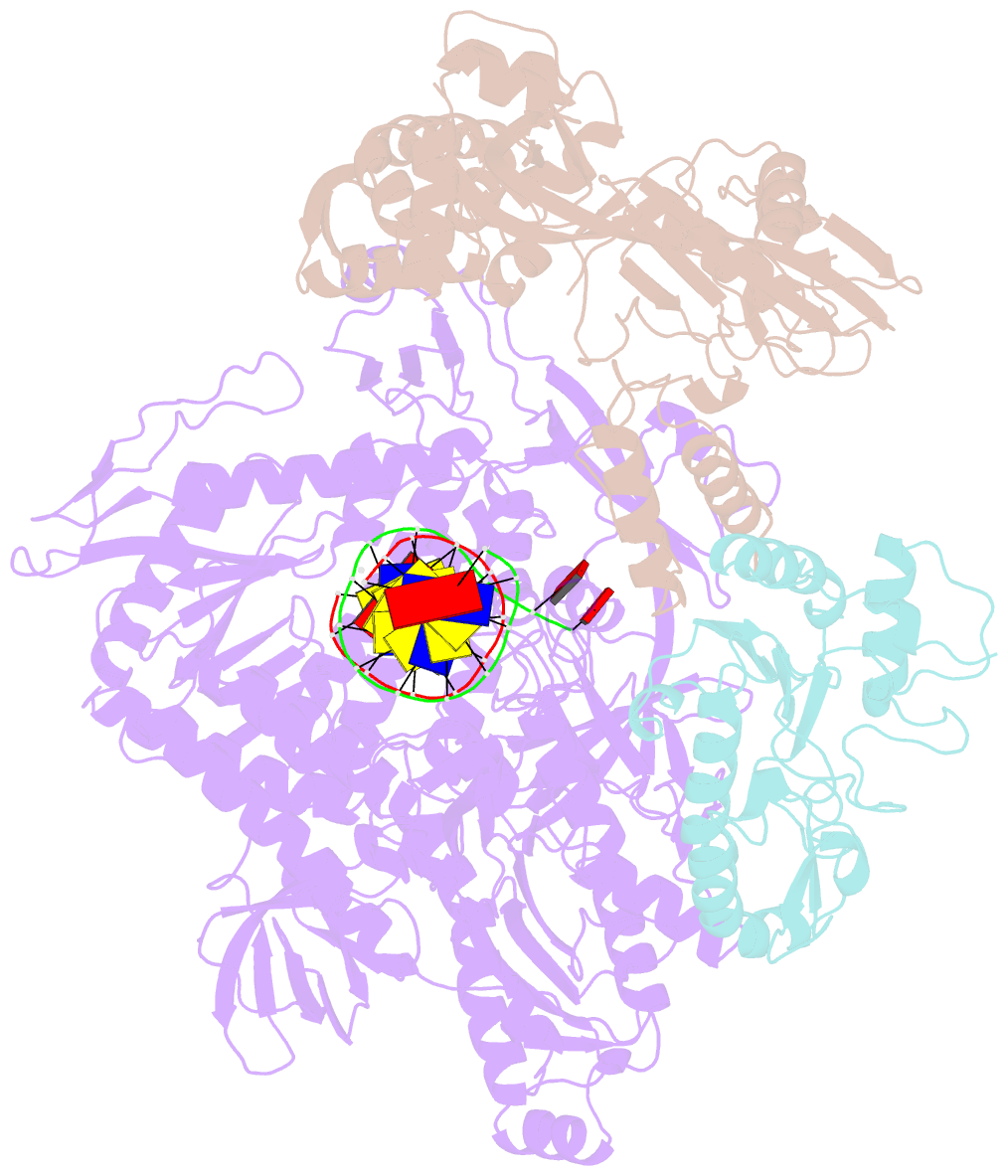
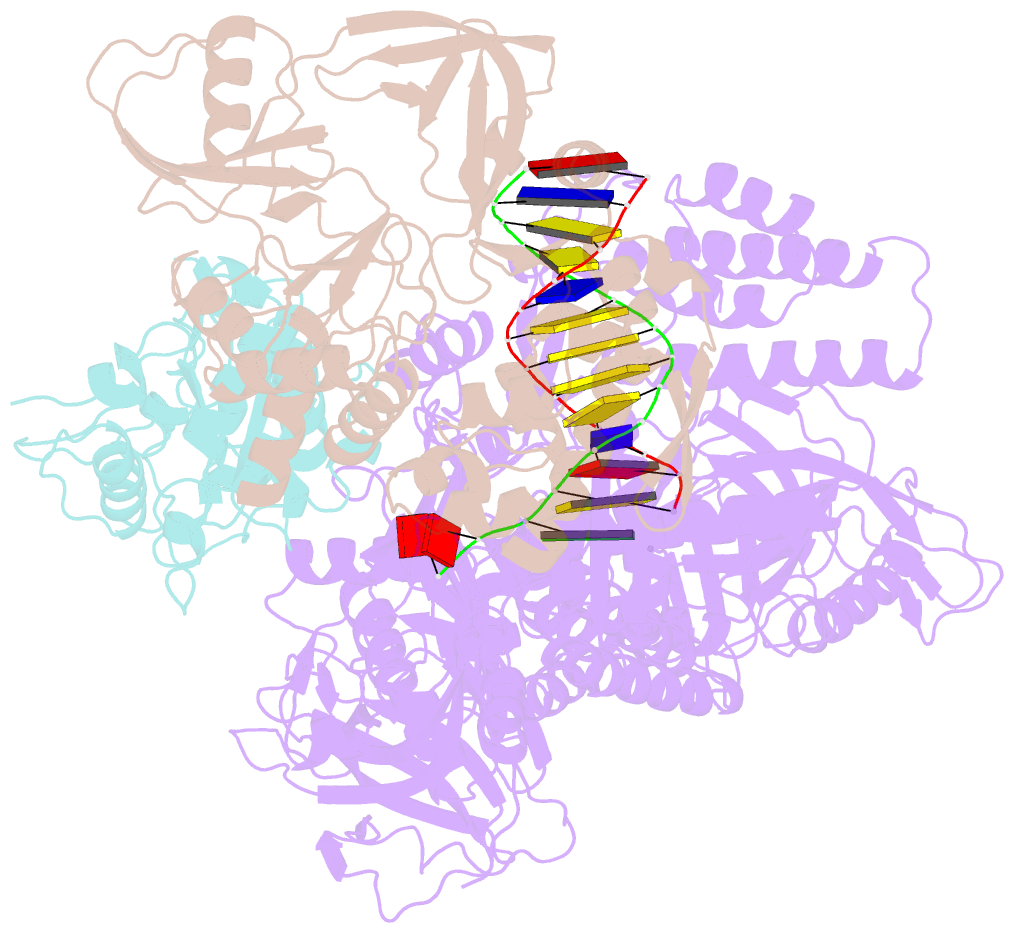
Summary information and primary citation
- PDB-id
- 8k8s; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein-DNA
- Method
- cryo-EM (3.06 Å)
- Summary
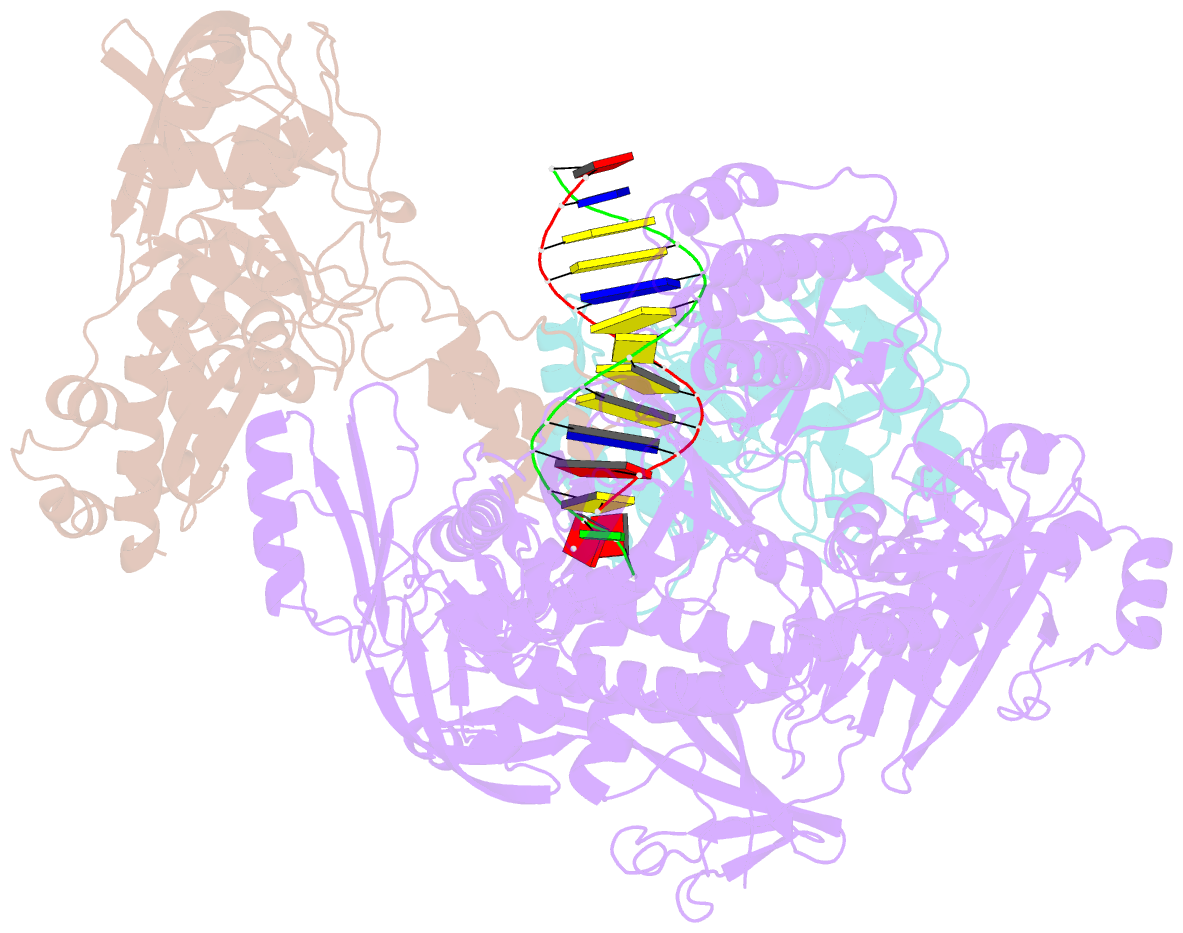
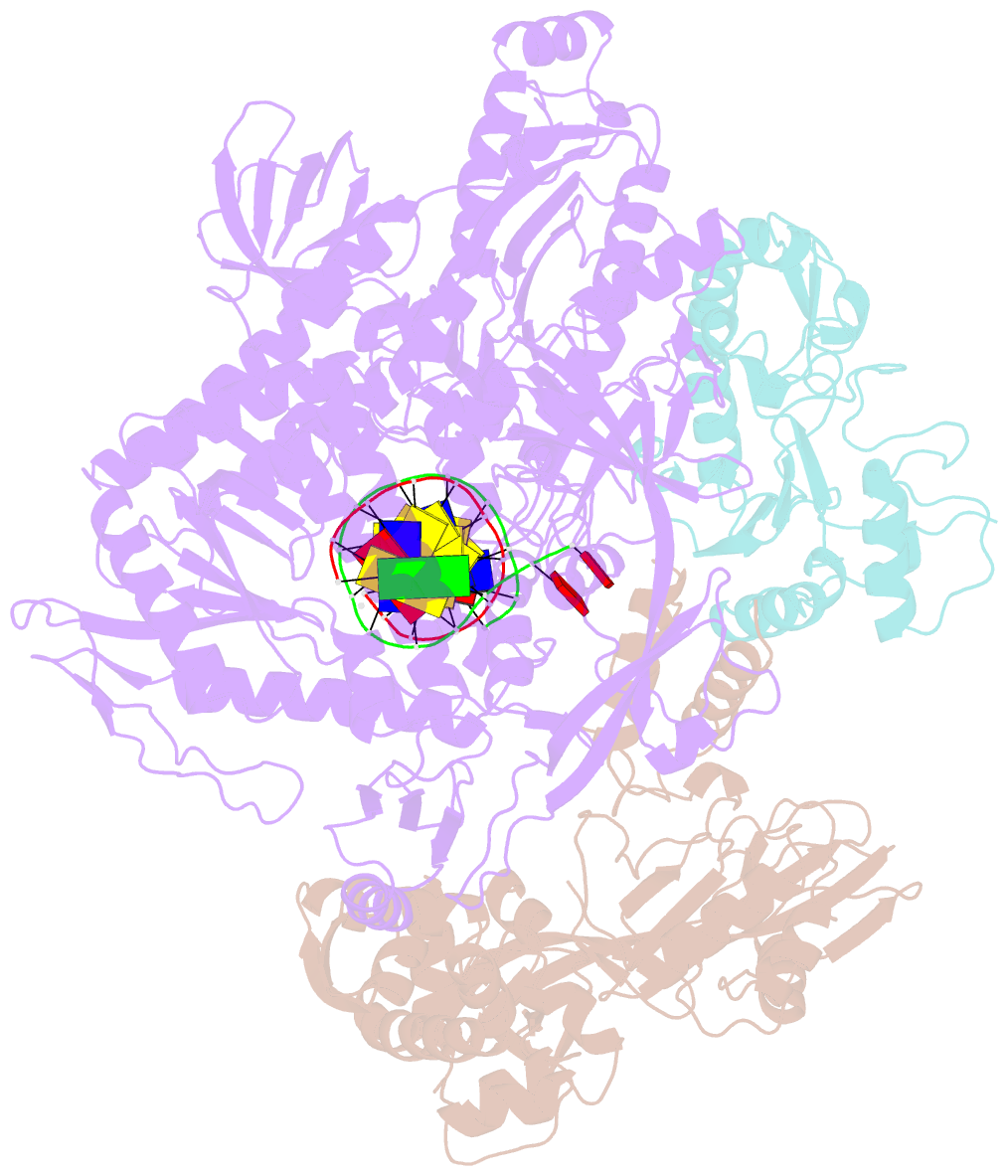
- F8-a22-e4 complex of mpxv in complex with DNA and ara-ctp
- Reference
- Shen Y, Li Y, Yan R (2024): "Structural basis for the inhibition mechanism of the DNA polymerase holoenzyme from mpox virus." Structure, 32, 654-661.e3. doi: 10.1016/j.str.2024.03.004.
- Abstract
- There are three key components at the core of the mpox virus (MPXV) DNA polymerase holoenzyme: DNA polymerase F8, processivity factors A22, and the Uracil-DNA glycosylase E4. The holoenzyme is recognized as a vital antiviral target because MPXV replicates in the cytoplasm of host cells. Nucleotide analogs such as cidofovir and cytarabine (Ara-C) have shown potential in curbing MPXV replication and they also display promise against other poxviruses. However, the mechanism behind their inhibitory effects remains unclear. Here, we present the cryo-EM structure of the DNA polymerase holoenzyme F8/A22/E4 bound with its competitive inhibitor Ara-C-derived cytarabine triphosphate (Ara-CTP) at an overall resolution of 3.0 Å and reveal its inhibition mechanism. Ara-CTP functions as a direct chain terminator in proximity to the deoxycytidine triphosphate (dCTP)-binding site. The extra hydrogen bond formed with Asn665 makes it more potent in binding than dCTP. Asn665 is conserved among eukaryotic B-family polymerases.