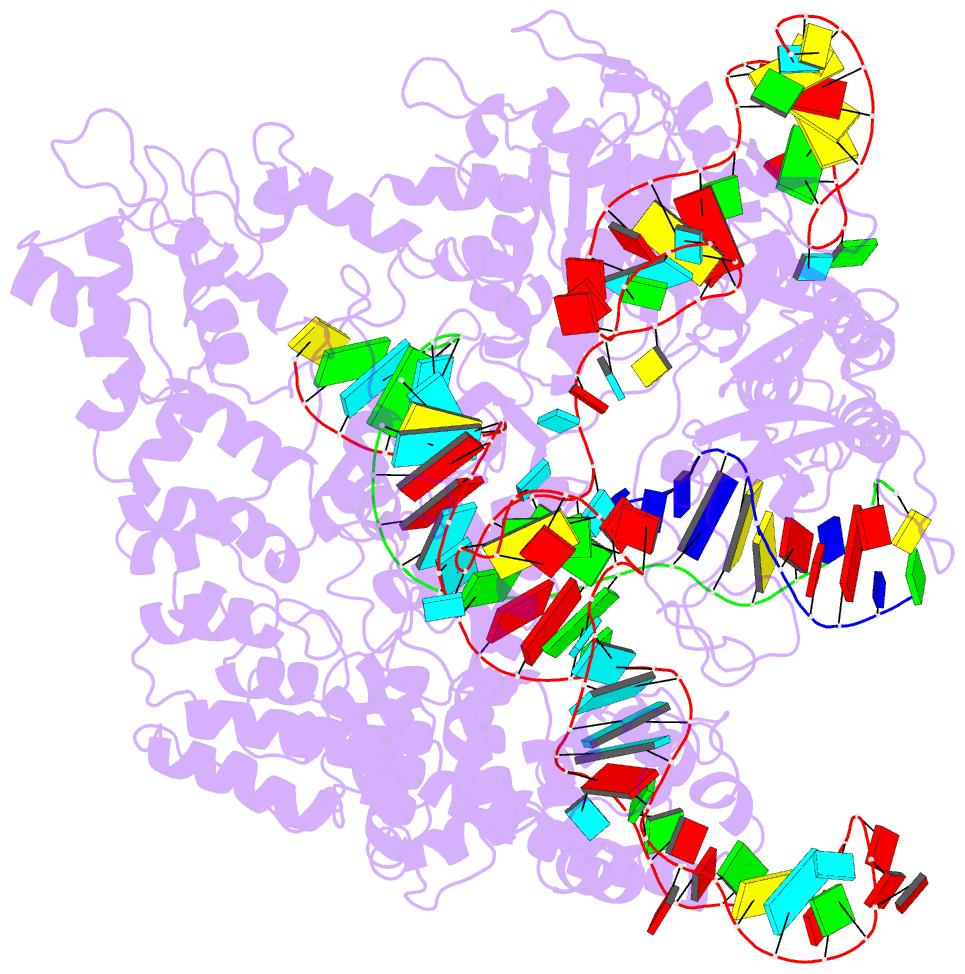
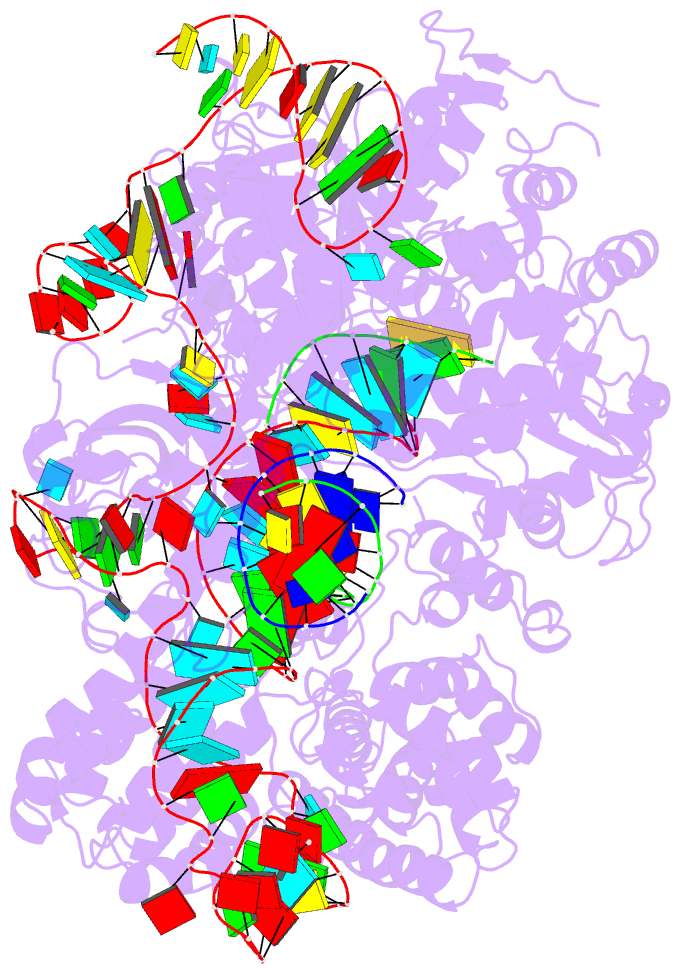
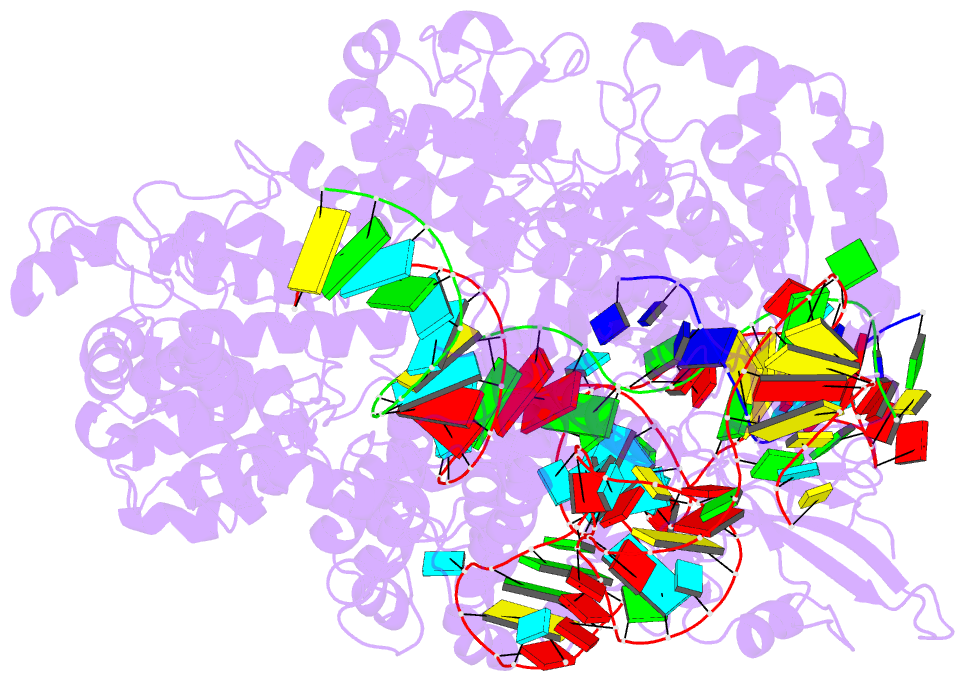
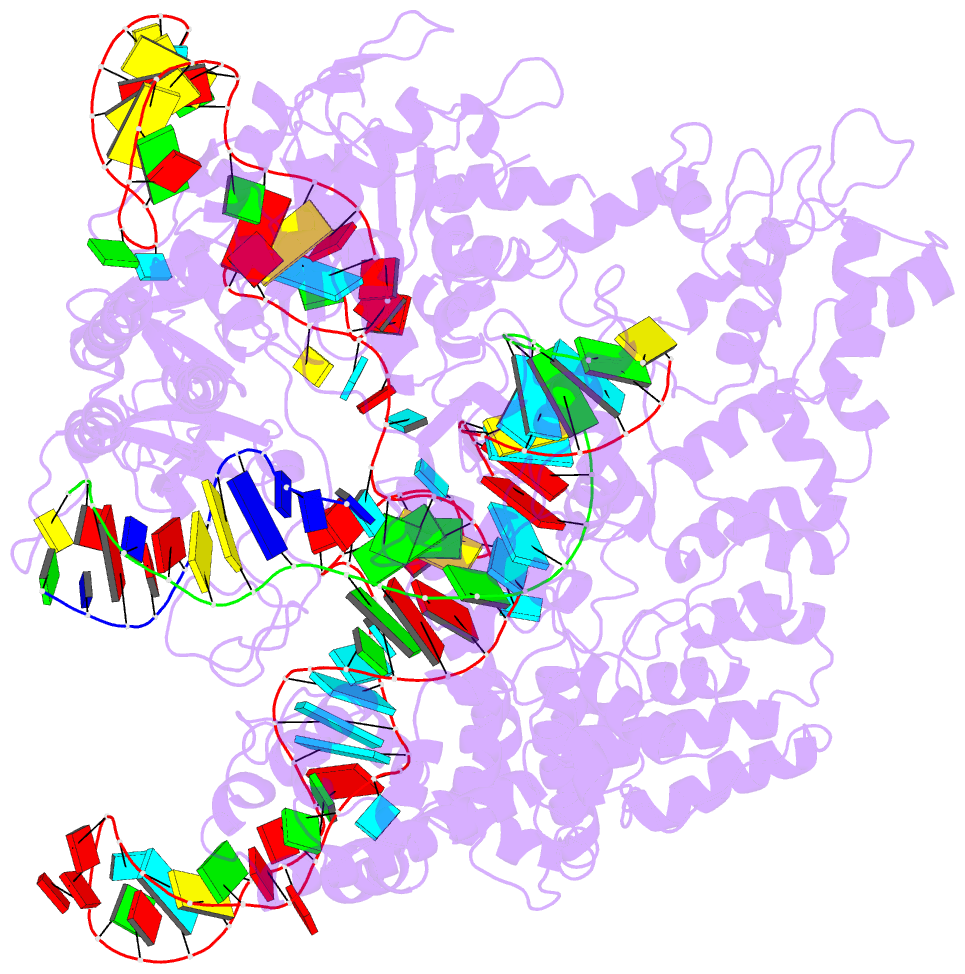
Summary information and primary citation
- PDB-id
- 8kam; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA-DNA
- Method
- X-ray (3.91 Å)
- Summary
- Crystal structure of spycas9 in complex with sgrna and 16nt target DNA
- Reference
- Chen J, Chen Y, Huang L, Lin X, Chen H, Xiang W, Liu L (2024): "Trans-nuclease activity of Cas9 activated by DNA or RNA target binding." Nat.Biotechnol. doi: 10.1038/s41587-024-02255-7.
- Abstract
- Type V and type VI CRISPR-Cas systems have been shown to cleave nonspecific single-stranded DNA (ssDNA) or single-stranded RNA (ssRNA) in trans, but this has not been observed in type II CRISPR-Cas systems using single guide RNA. We show here that the type II CRISPR-Cas9 systems directed by CRISPR RNA and trans-activating CRISPR RNA dual RNAs show RuvC domain-dependent trans-cleavage activity for both ssDNA and ssRNA substrates. Cas9 possesses sequence preferences for trans-cleavage substrates, preferring to cleave T- or C-rich ssDNA substrates. We find that the trans-cleavage activity of Cas9 can be activated by target ssDNA, double-stranded DNA and ssRNA. The crystal structure of Cas9 in complex with guide RNA and target RNA provides a structural basis for the binding of target RNA to activate Cas9. Based on the trans-cleavage activity of Cas9 and nucleic acid amplification technology, we develop the nucleic acid detection platforms DNA-activated Cas9 detection and RNA-activated Cas9 detection, which are capable of detecting DNA and RNA samples with high sensitivity and specificity.