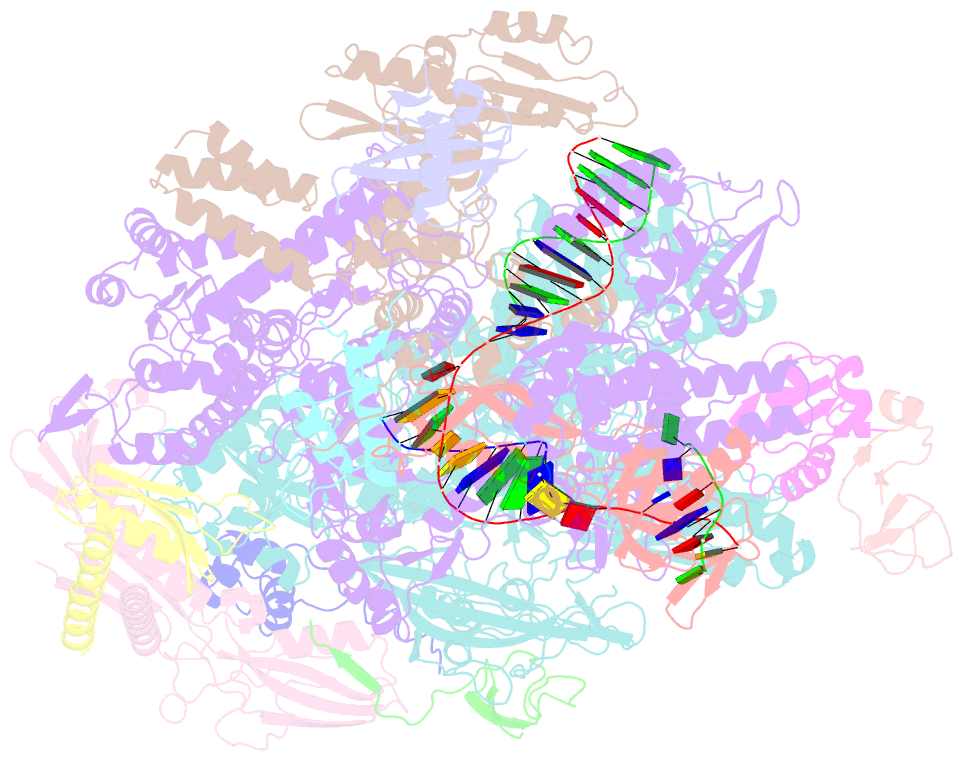
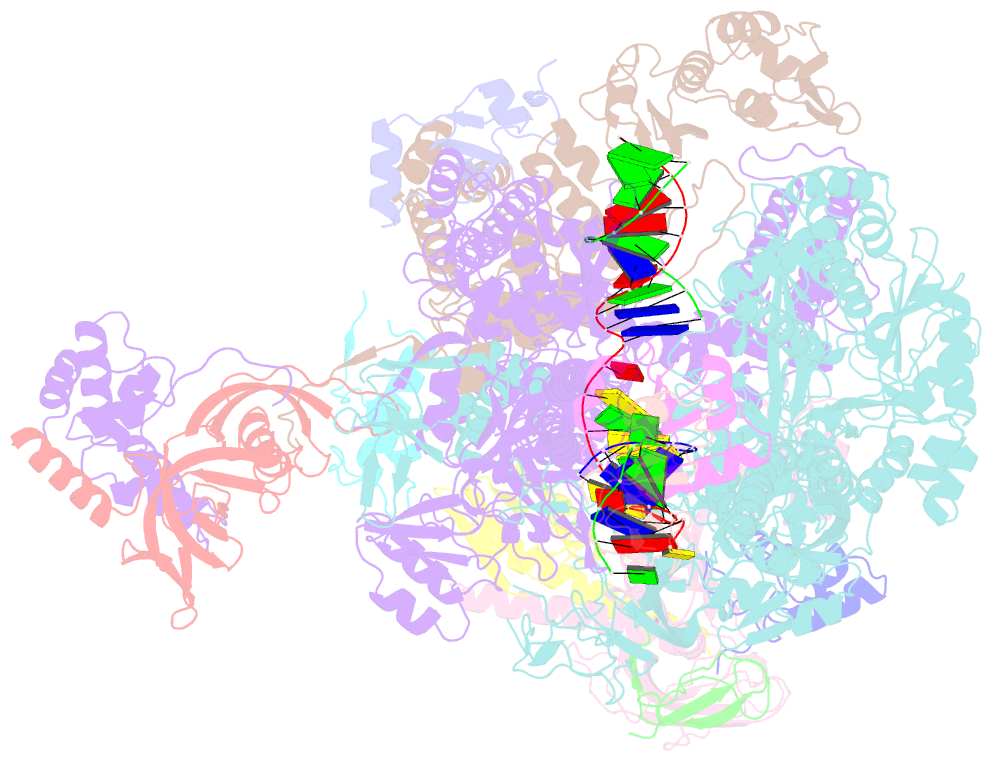
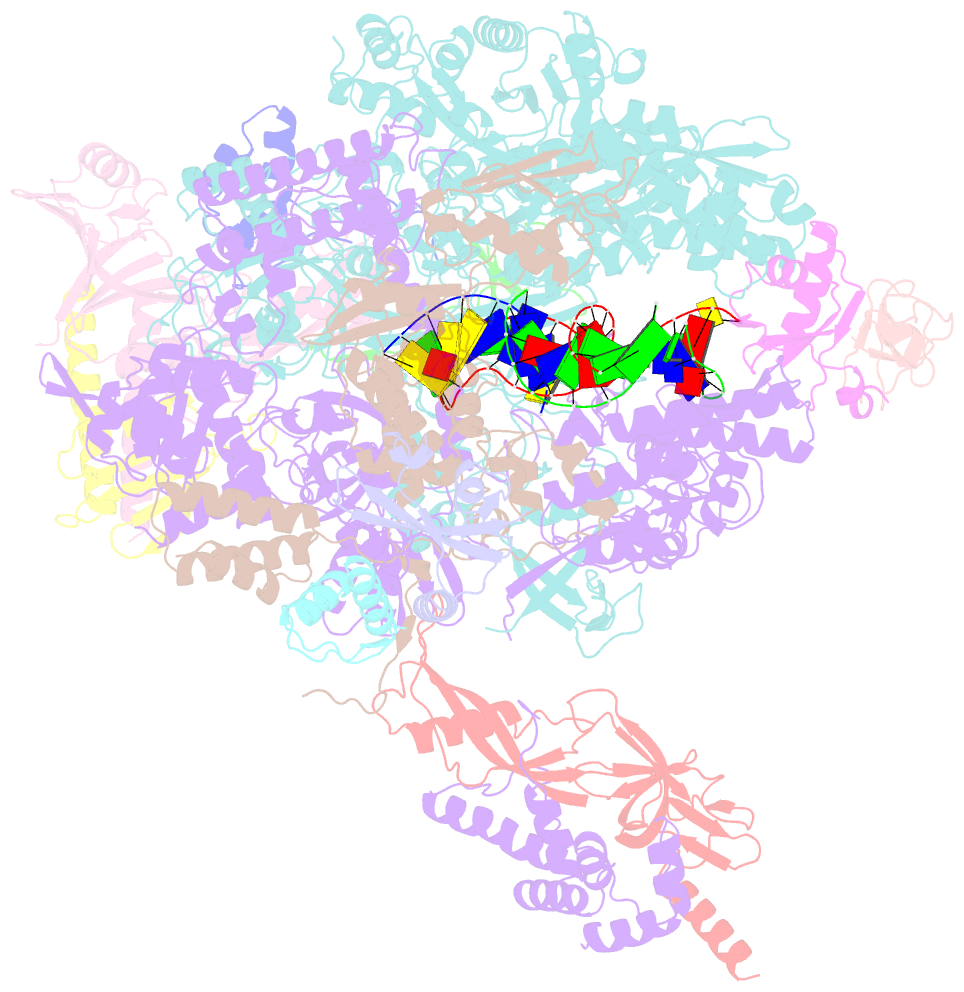
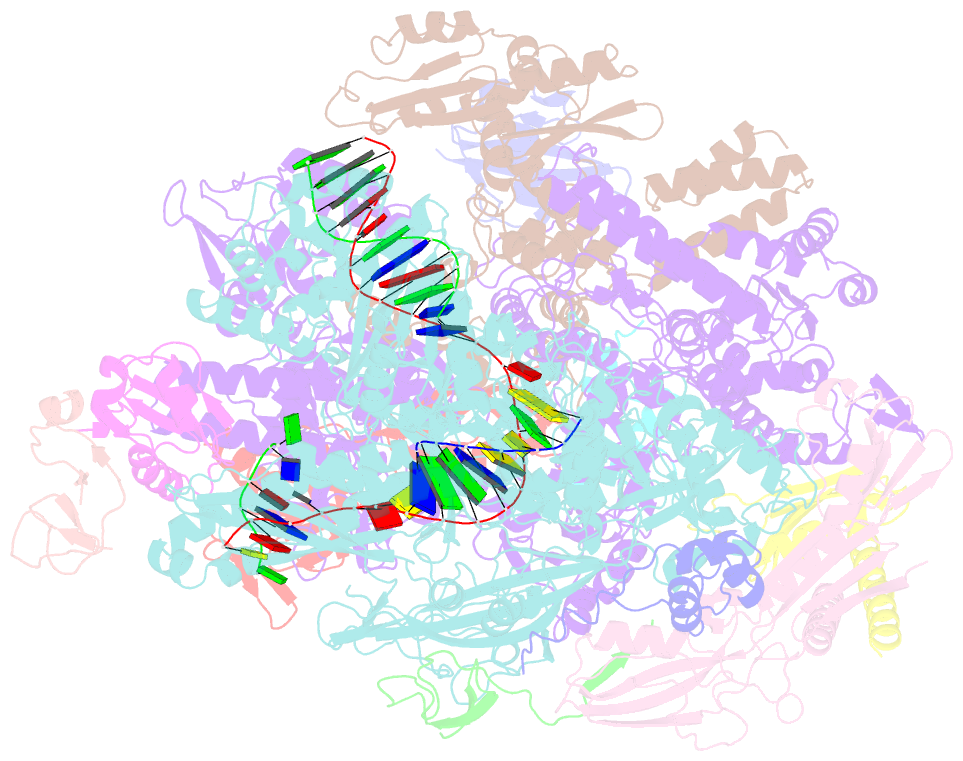
Summary information and primary citation
- PDB-id
- 8oki; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- cryo-EM (3.45 Å)
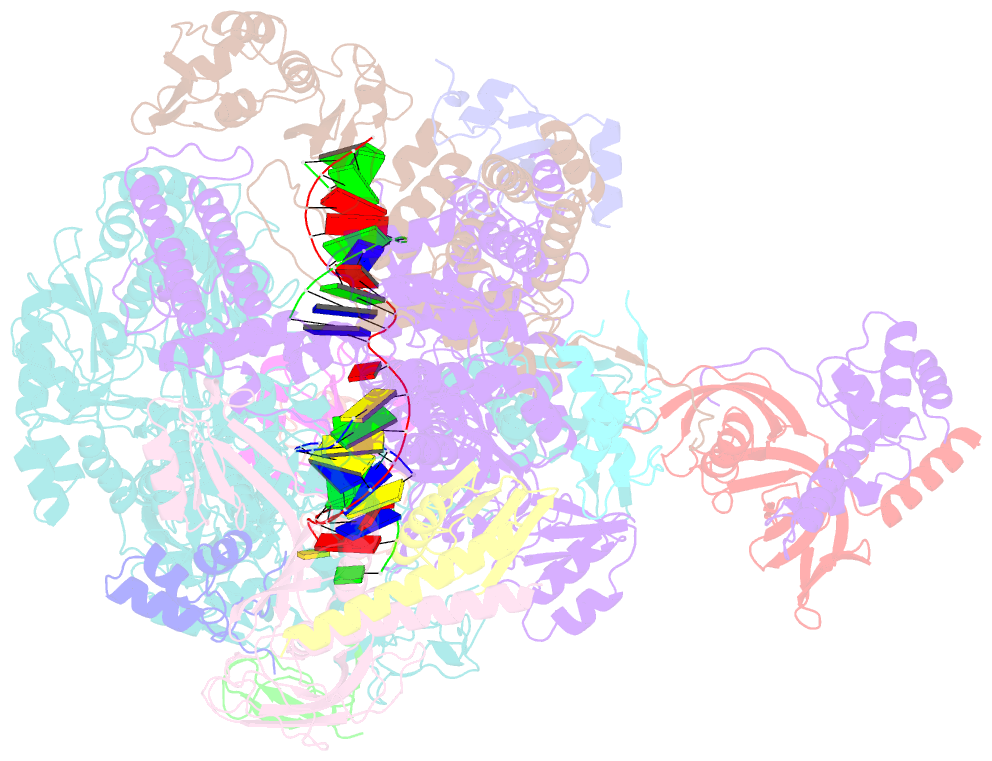
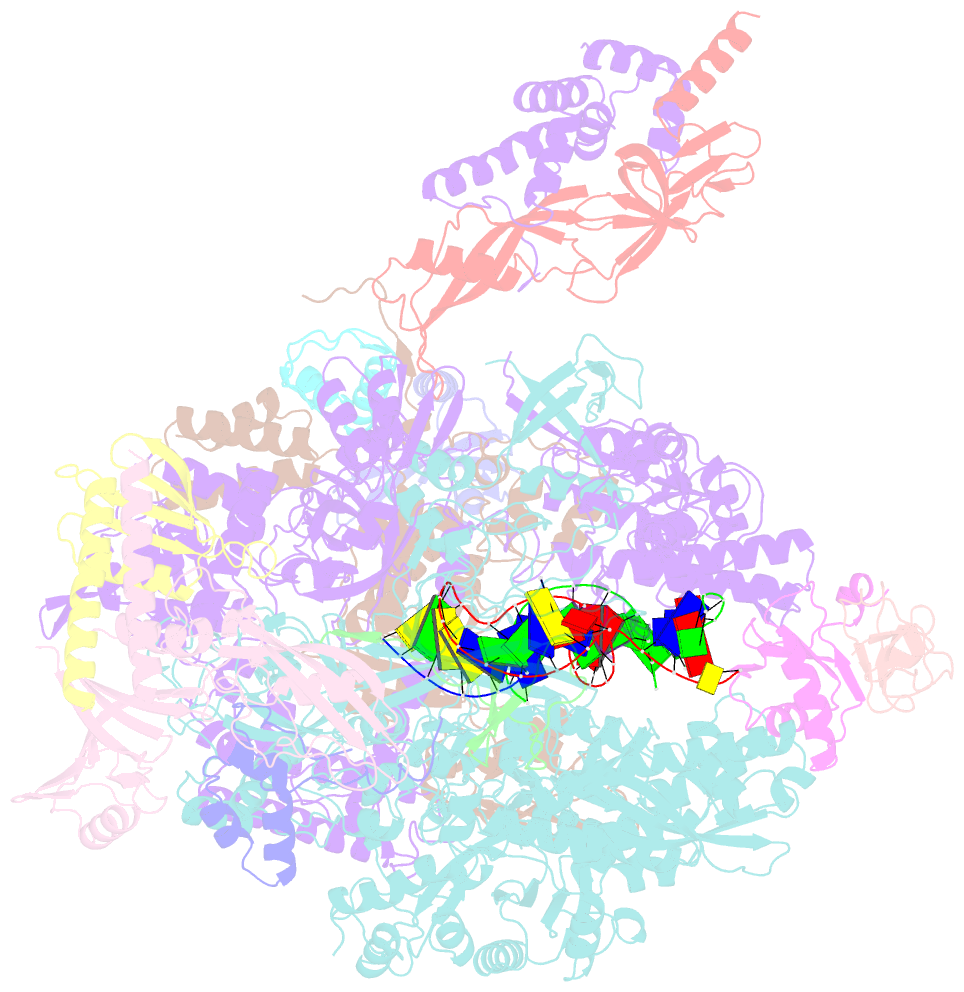
- Summary
- cryo-EM structure of pyrococcus furiosus transcription elongation complex bound to spt4-5
- Reference
- Tarau D, Grunberger F, Pilsl M, Reichelt R, Heiss F, Konig S, Urlaub H, Hausner W, Engel C, Grohmann D (2024): "Structural basis of archaeal RNA polymerase transcription elongation and Spt4/5 recruitment." Nucleic Acids Res., 52, 6017-6035. doi: 10.1093/nar/gkae282.
- Abstract
- Archaeal transcription is carried out by a multi-subunit RNA polymerase (RNAP) that is highly homologous in structure and function to eukaryotic RNAP II. Among the set of basal transcription factors, only Spt5 is found in all domains of life, but Spt5 has been shaped during evolution, which is also reflected in the heterodimerization of Spt5 with Spt4 in Archaea and Eukaryotes. To unravel the mechanistic basis of Spt4/5 function in Archaea, we performed structure-function analyses using the archaeal transcriptional machinery of Pyrococcus furiosus (Pfu). We report single-particle cryo-electron microscopy reconstructions of apo RNAP and the archaeal elongation complex (EC) in the absence and presence of Spt4/5. Surprisingly, Pfu Spt4/5 also binds the RNAP in the absence of nucleic acids in a distinct super-contracted conformation. We show that the RNAP clamp/stalk module exhibits conformational flexibility in the apo state of RNAP and that the enzyme contracts upon EC formation or Spt4/5 engagement. We furthermore identified a contact of the Spt5-NGN domain with the DNA duplex that stabilizes the upstream boundary of the transcription bubble and impacts Spt4/5 activity in vitro. This study, therefore, provides the structural basis for Spt4/5 function in archaeal transcription and reveals a potential role beyond the well-described support of elongation.