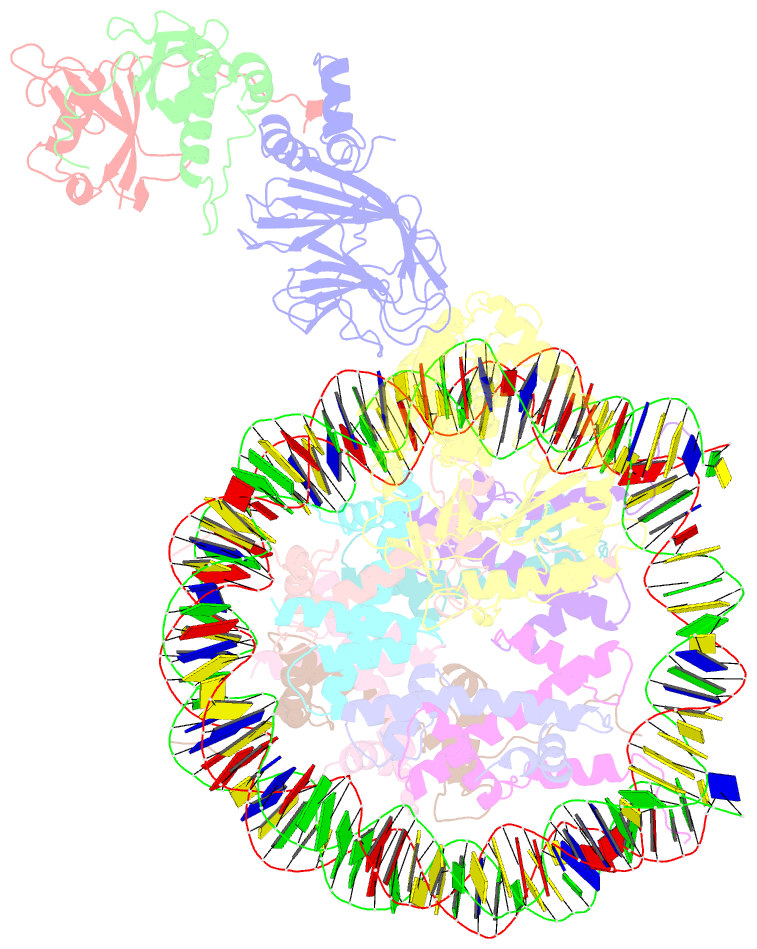
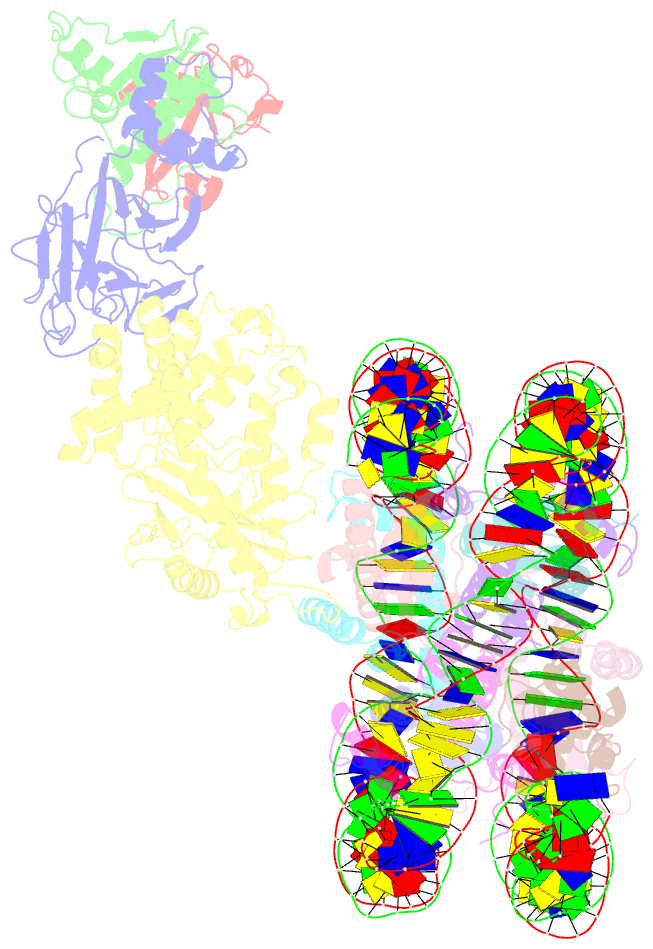
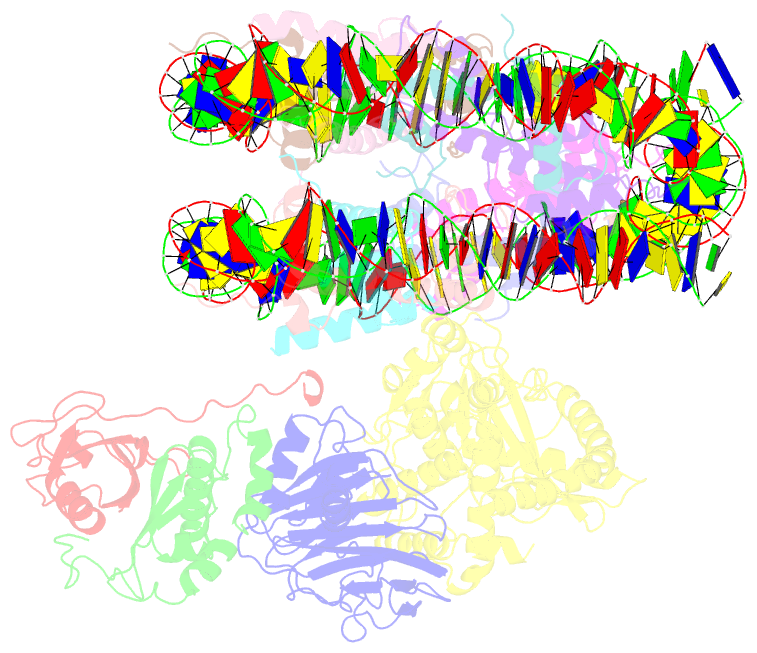
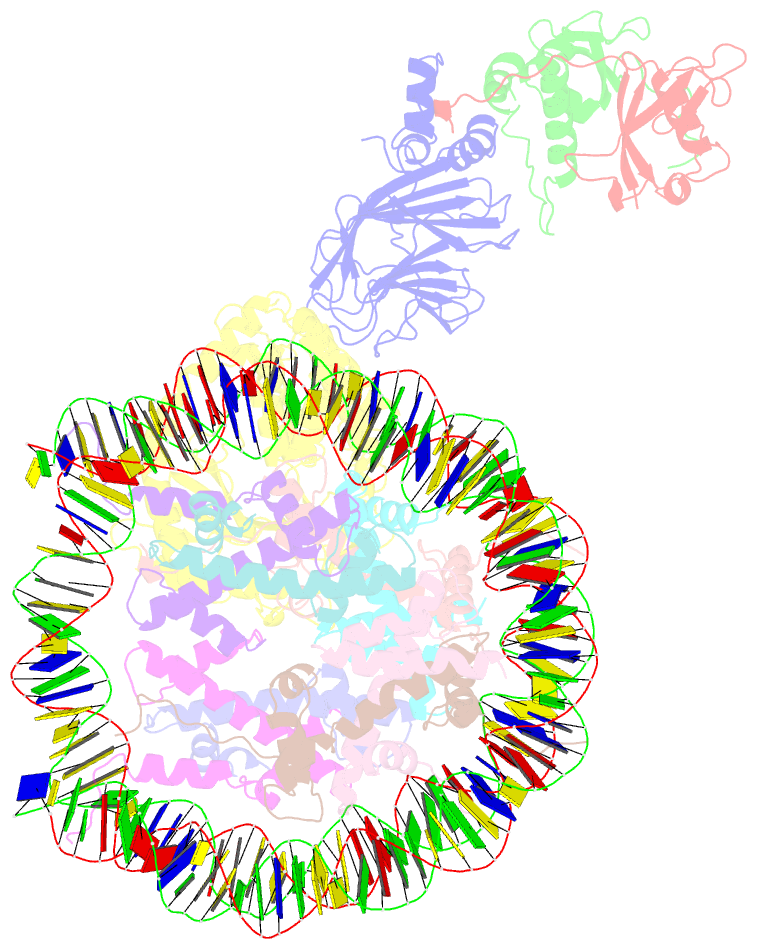
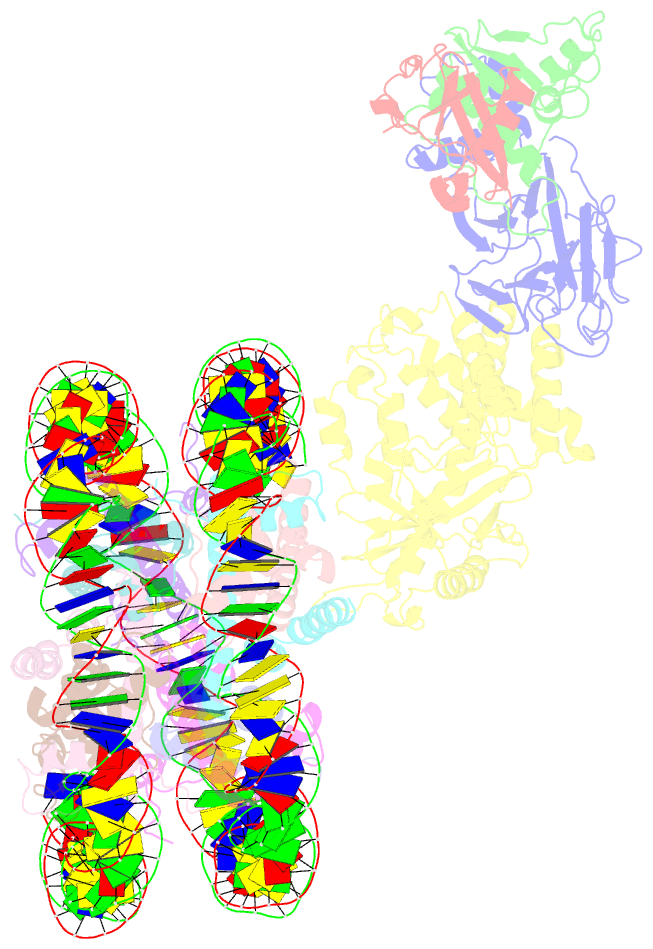
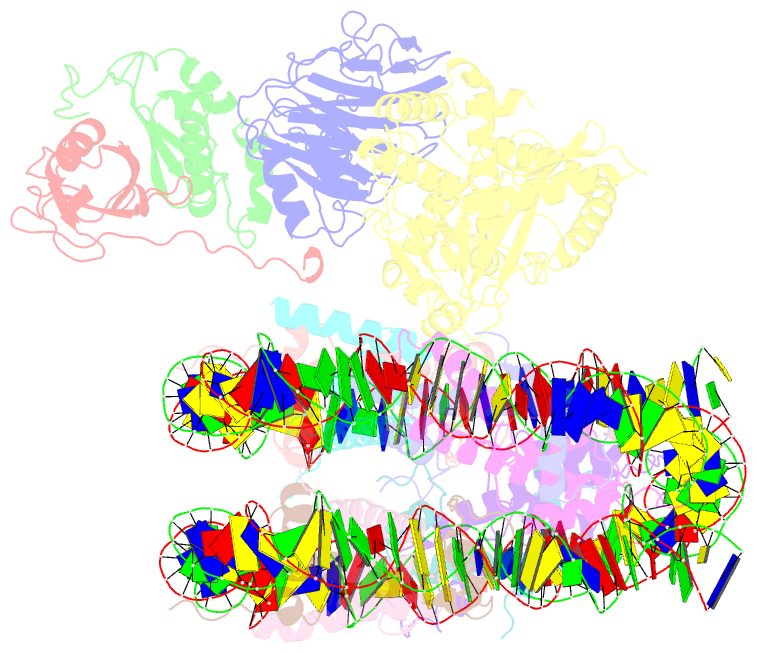
Summary information and primary citation
- PDB-id
- 8ol1; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- immune system
- Method
- cryo-EM (3.5 Å)
- Summary
- Cgas-nucleosome in complex with spsb3-elobc (composite structure)
- Reference
- Xu P, Liu Y, Liu C, Guey B, Li L, Melenec P, Ricci J, Ablasser A (2024): "The CRL5-SPSB3 ubiquitin ligase targets nuclear cGAS for degradation." Nature, 627, 873-879. doi: 10.1038/s41586-024-07112-w.
- Abstract
- Cyclic GMP-AMP synthase (cGAS) senses aberrant DNA during infection, cancer and inflammatory disease, and initiates potent innate immune responses through the synthesis of 2'3'-cyclic GMP-AMP (cGAMP)1-7. The indiscriminate activity of cGAS towards DNA demands tight regulatory mechanisms that are necessary to maintain cell and tissue homeostasis under normal conditions. Inside the cell nucleus, anchoring to nucleosomes and competition with chromatin architectural proteins jointly prohibit cGAS activation by genomic DNA8-15. However, the fate of nuclear cGAS and its role in cell physiology remains unclear. Here we show that the ubiquitin proteasomal system (UPS) degrades nuclear cGAS in cycling cells. We identify SPSB3 as the cGAS-targeting substrate receptor that associates with the cullin-RING ubiquitin ligase 5 (CRL5) complex to ligate ubiquitin onto nuclear cGAS. A cryo-electron microscopy structure of nucleosome-bound cGAS in a complex with SPSB3 reveals a highly conserved Asn-Asn (NN) minimal degron motif at the C terminus of cGAS that directs SPSB3 recruitment, ubiquitylation and cGAS protein stability. Interference with SPSB3-regulated nuclear cGAS degradation primes cells for type I interferon signalling, conferring heightened protection against infection by DNA viruses. Our research defines protein degradation as a determinant of cGAS regulation in the nucleus and provides structural insights into an element of cGAS that is amenable to therapeutic exploitation.