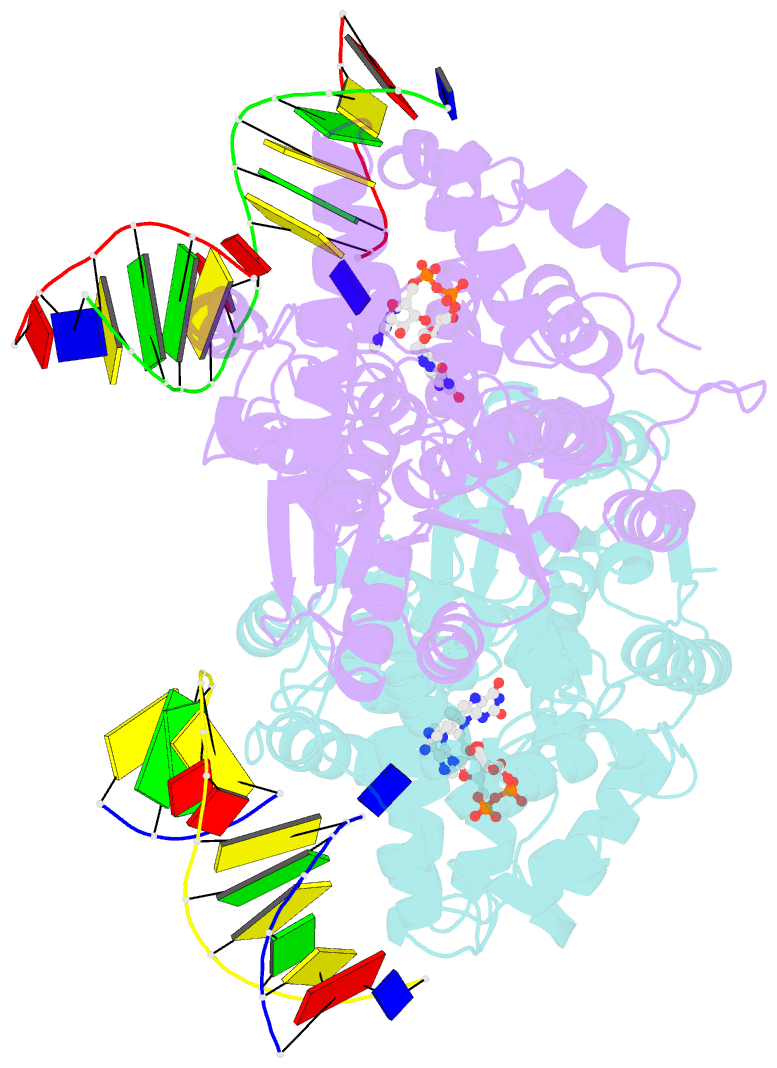
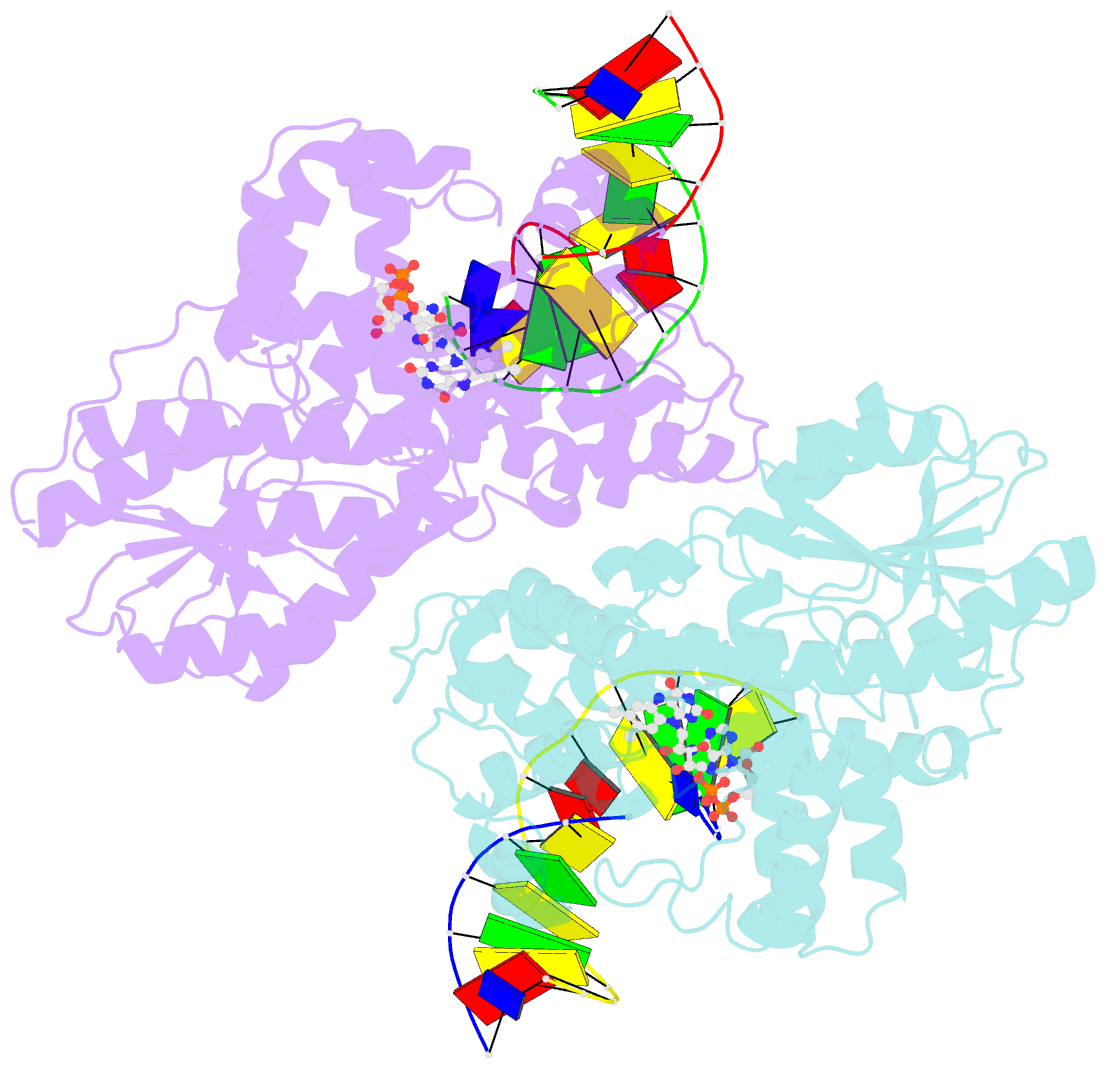
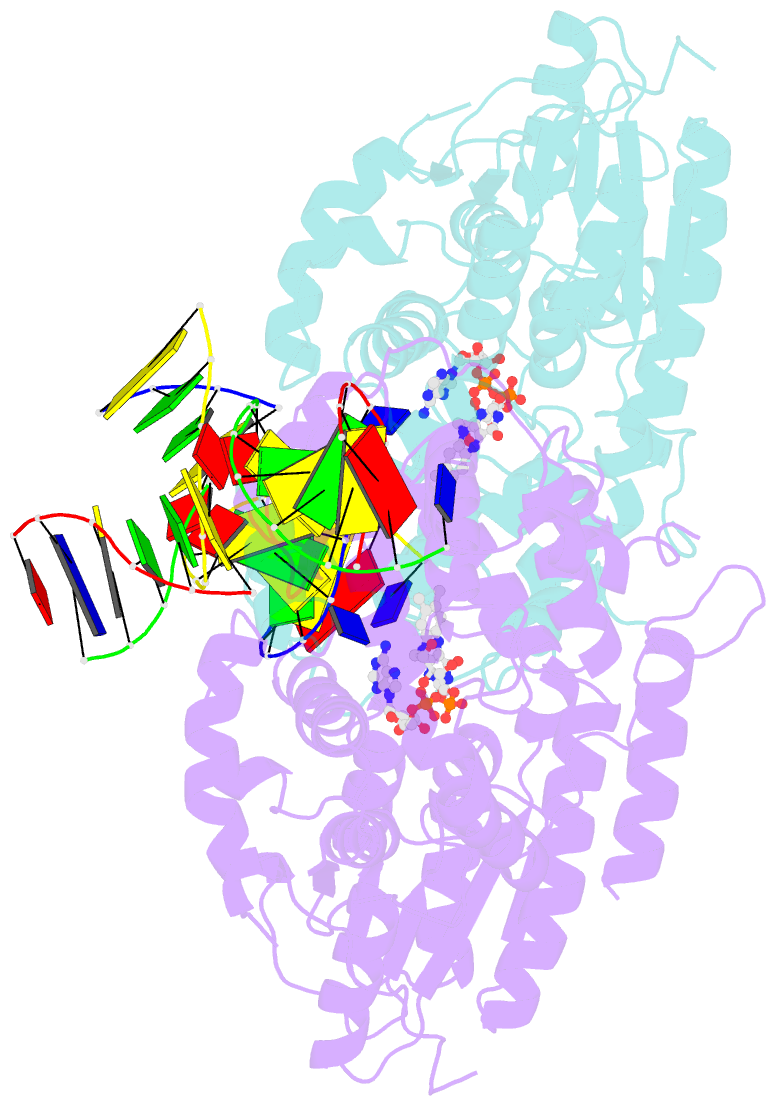
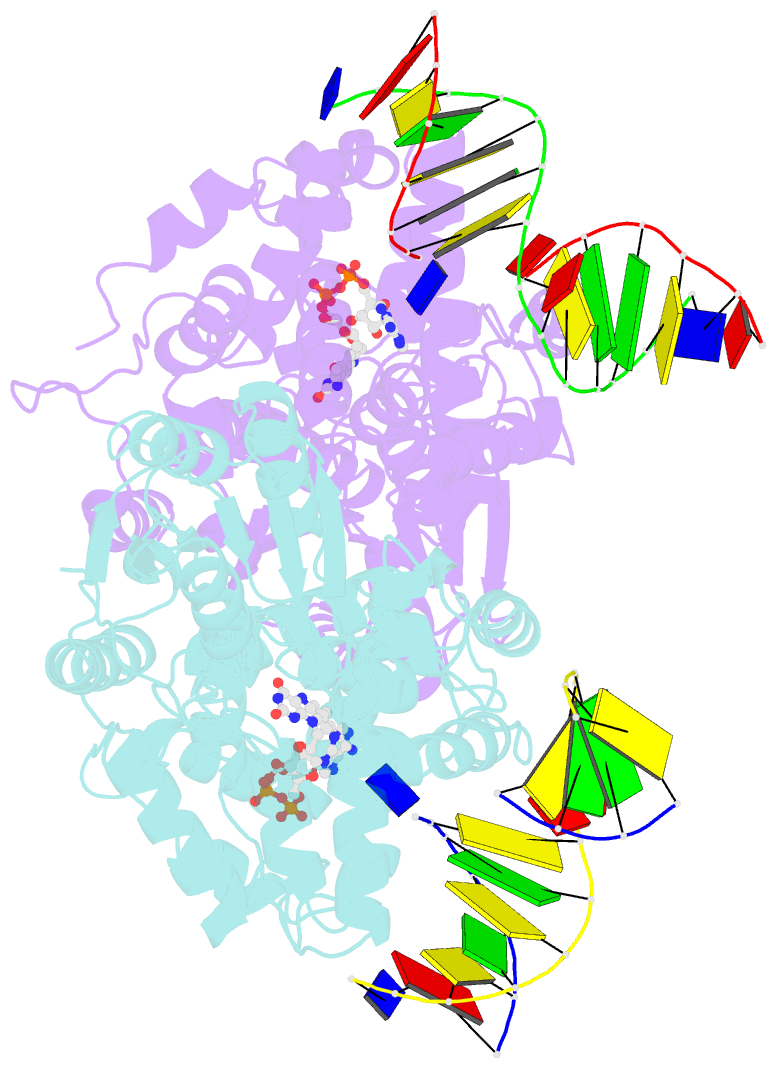
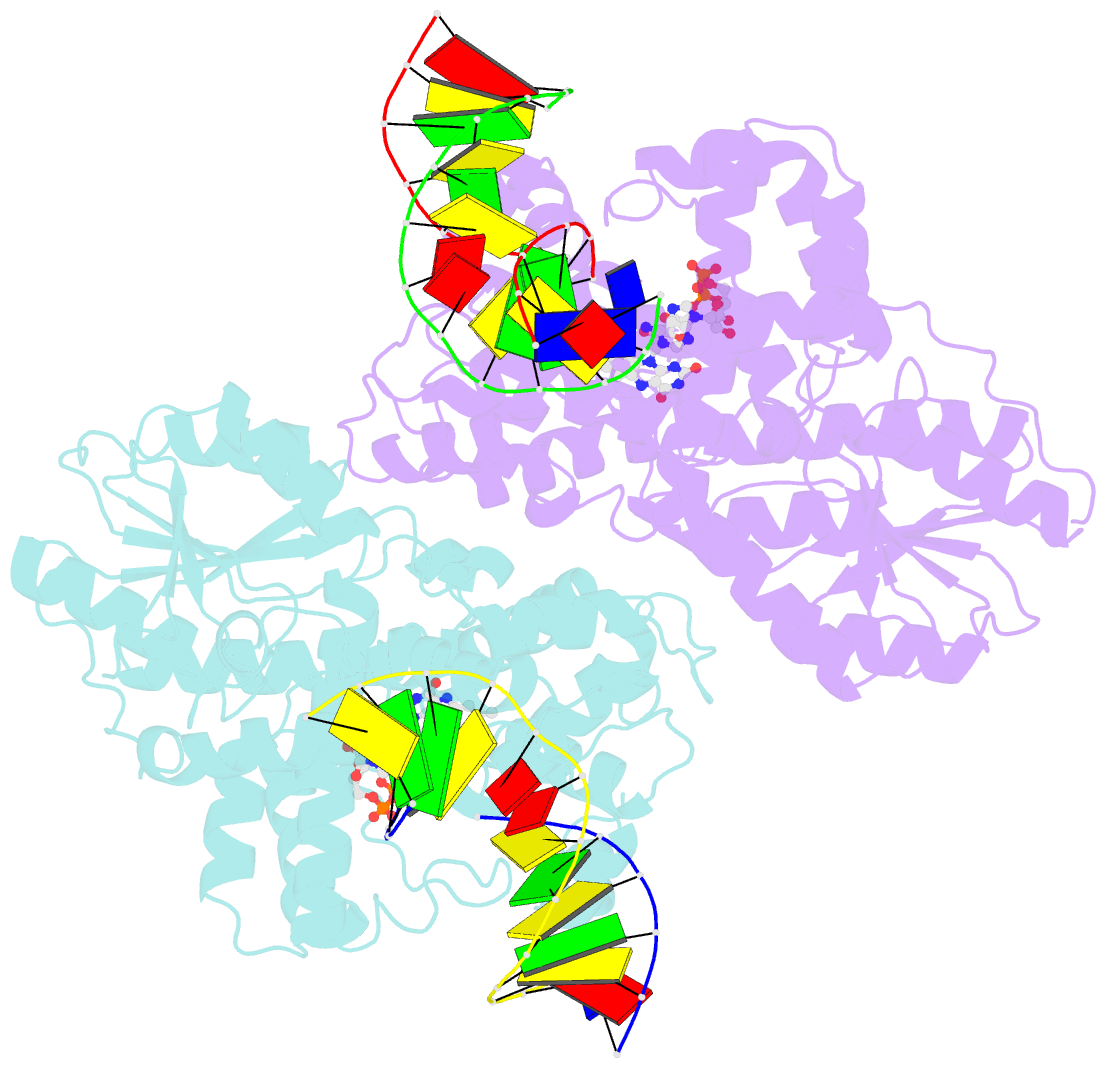
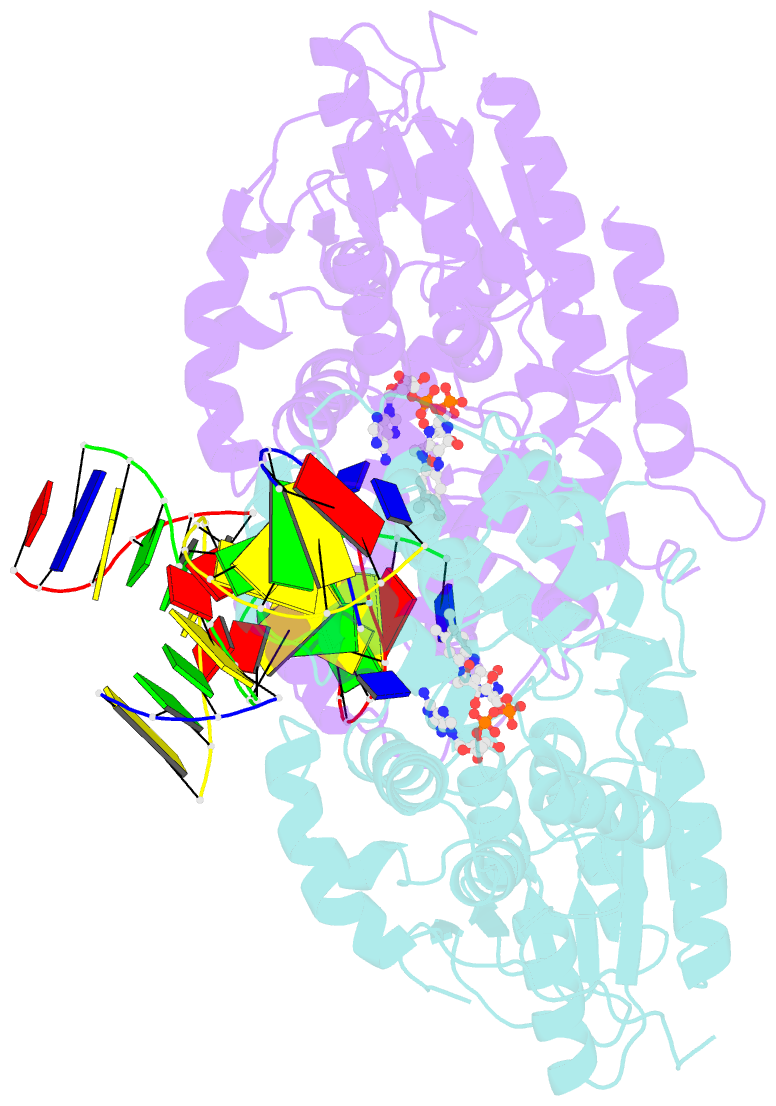
Summary information and primary citation
- PDB-id
- 8oy3; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- lyase
- Method
- X-ray (2.16 Å)
- Summary
- Time-resolved sfx structure of the class ii photolyase complexed with a thymine dimer (3 picosecond pump-probe delay)
- Reference
- Christou NE, Apostolopoulou V, Melo DVM, Ruppert M, Fadini A, Henkel A, Sprenger J, Oberthuer D, Gunther S, Pateras A, Rahmani Mashhour A, Yefanov OM, Galchenkova M, Reinke PYA, Kremling V, Scheer TES, Lange ER, Middendorf P, Schubert R, De Zitter E, Lumbao-Conradson K, Herrmann J, Rahighi S, Kunavar A, Beale EV, Beale JH, Cirelli C, Johnson PJM, Dworkowski F, Ozerov D, Bertrand Q, Wranik M, Bacellar C, Bajt S, Wakatsuki S, Sellberg JA, Huse N, Turk D, Chapman HN, Lane TJ (2023): "Time-resolved crystallography captures light-driven DNA repair." Science, 382, 1015-1020. doi: 10.1126/science.adj4270.
- Abstract
- Photolyase is an enzyme that uses light to catalyze DNA repair. To capture the reaction intermediates involved in the enzyme's catalytic cycle, we conducted a time-resolved crystallography experiment. We found that photolyase traps the excited state of the active cofactor, flavin adenine dinucleotide (FAD), in a highly bent geometry. This excited state performs electron transfer to damaged DNA, inducing repair. We show that the repair reaction, which involves the lysis of two covalent bonds, occurs through a single-bond intermediate. The transformation of the substrate into product crowds the active site and disrupts hydrogen bonds with the enzyme, resulting in stepwise product release, with the 3' thymine ejected first, followed by the 5' base.