Summary information and primary citation
- PDB-id
- 8p4f; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- cryo-EM (4.0 Å)
- Summary
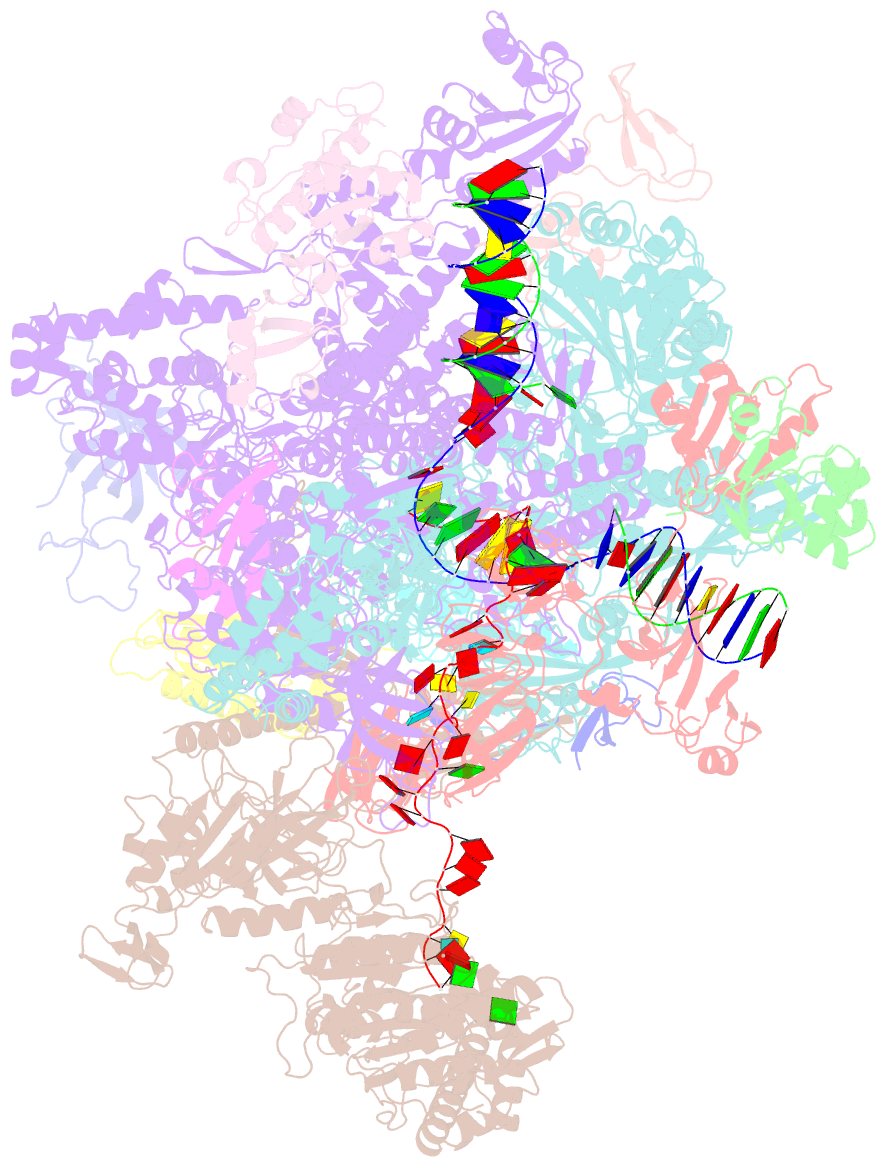
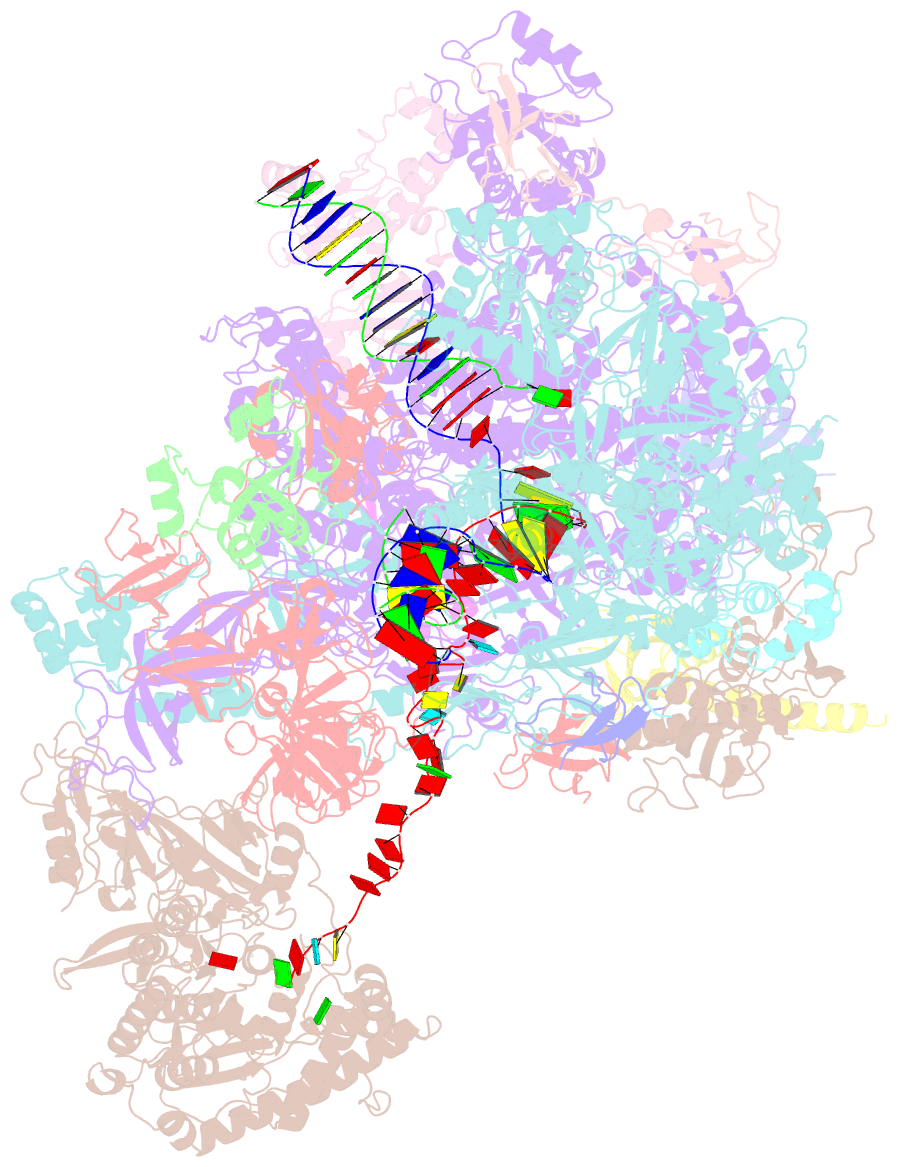
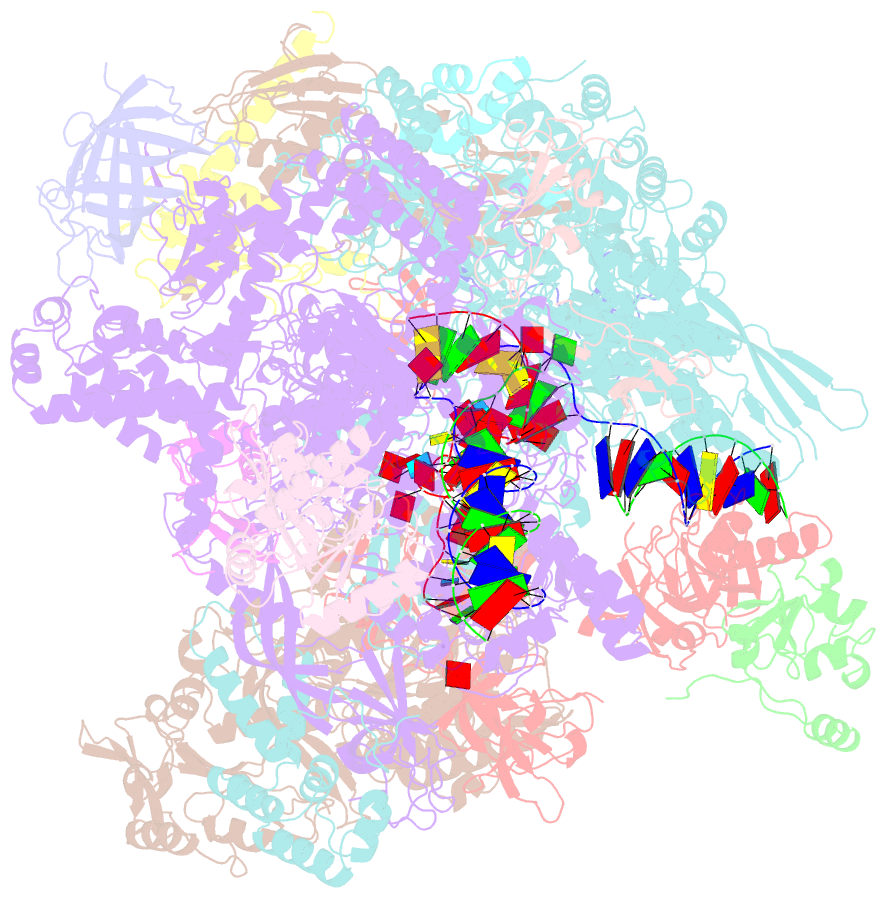
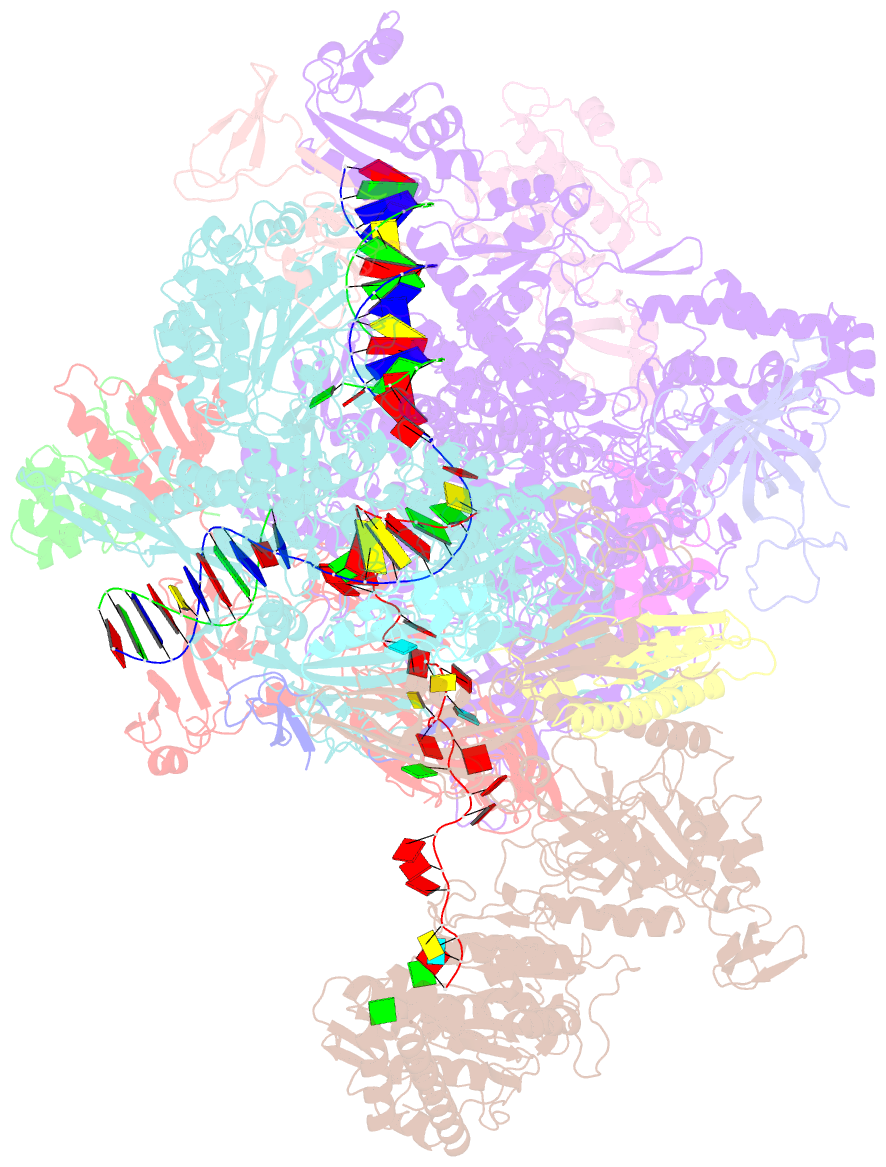
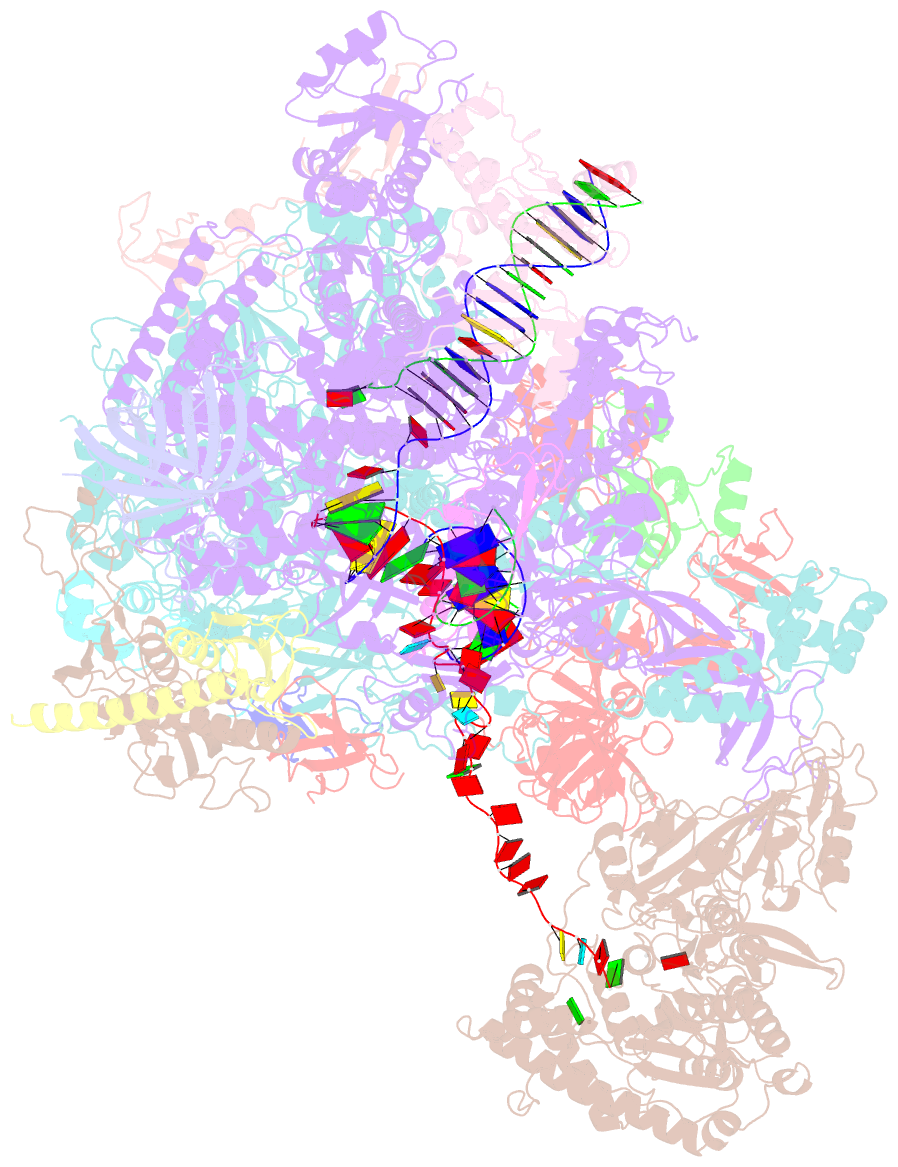
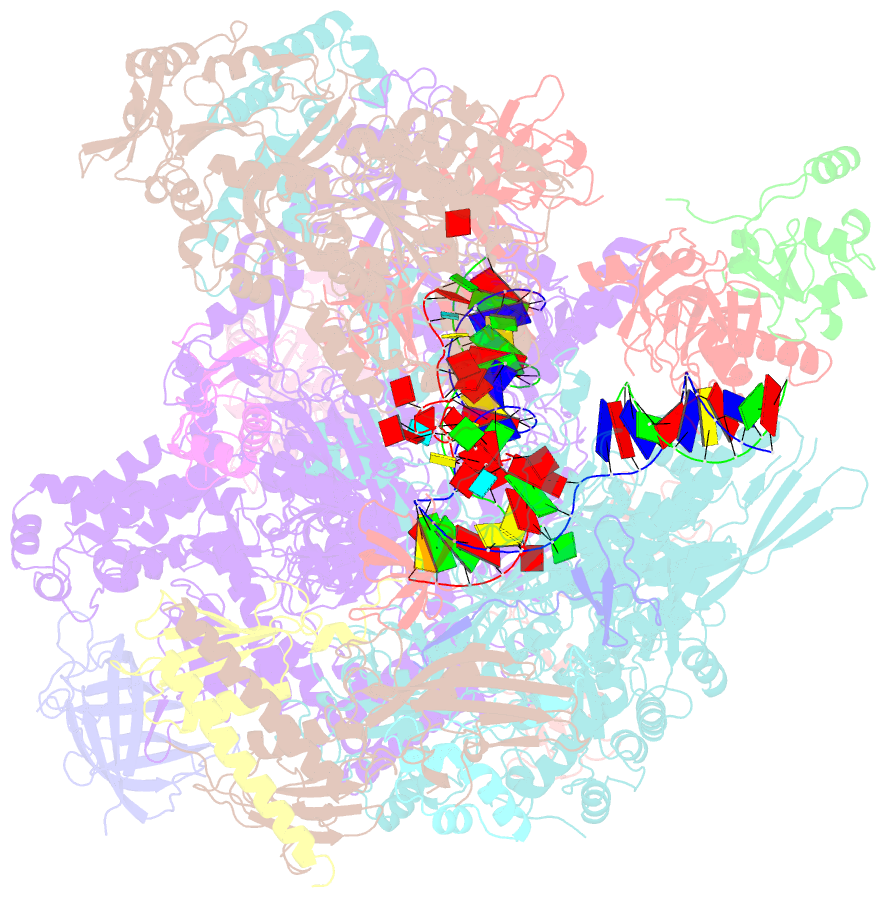
- Structural insights into human co-transcriptional capping - structure 6
- Reference
- Garg G, Dienemann C, Farnung L, Schwarz J, Linden A, Urlaub H, Cramer P (2023): "Structural insights into human co-transcriptional capping." Mol.Cell, 83, 2464. doi: 10.1016/j.molcel.2023.06.002.
- Abstract
- Co-transcriptional capping of the nascent pre-mRNA 5' end prevents degradation of RNA polymerase (Pol) II transcripts and suppresses the innate immune response. Here, we provide mechanistic insights into the three major steps of human co-transcriptional pre-mRNA capping based on six different cryoelectron microscopy (cryo-EM) structures. The human mRNA capping enzyme, RNGTT, first docks to the Pol II stalk to position its triphosphatase domain near the RNA exit site. The capping enzyme then moves onto the Pol II surface, and its guanylyltransferase receives the pre-mRNA 5'-diphosphate end. Addition of a GMP moiety can occur when the RNA is ∼22 nt long, sufficient to reach the active site of the guanylyltransferase. For subsequent cap(1) methylation, the methyltransferase CMTR1 binds the Pol II stalk and can receive RNA after it is grown to ∼29 nt in length. The observed rearrangements of capping factors on the Pol II surface may be triggered by the completion of catalytic reaction steps and are accommodated by domain movements in the elongation factor DRB sensitivity-inducing factor (DSIF).