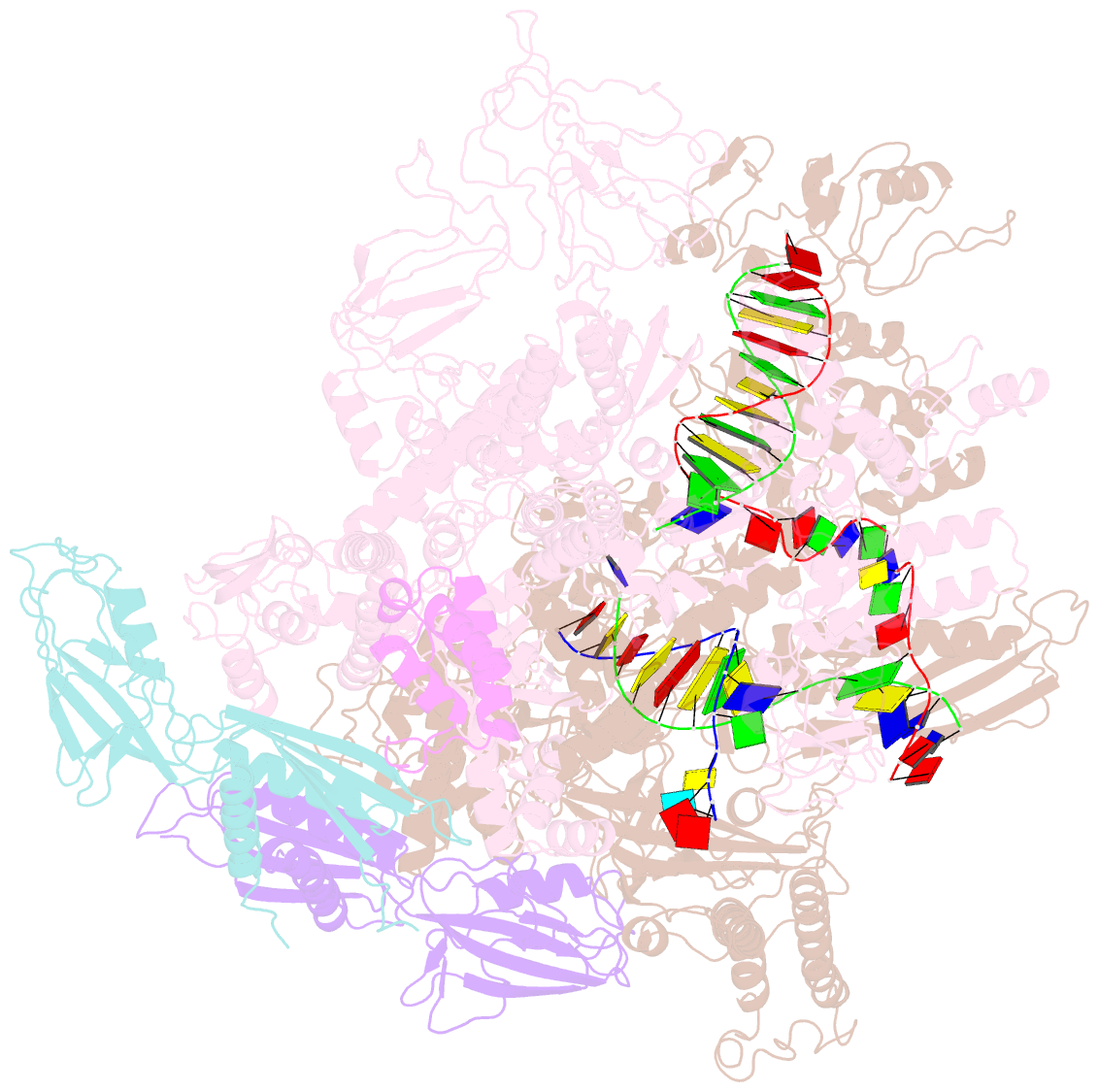
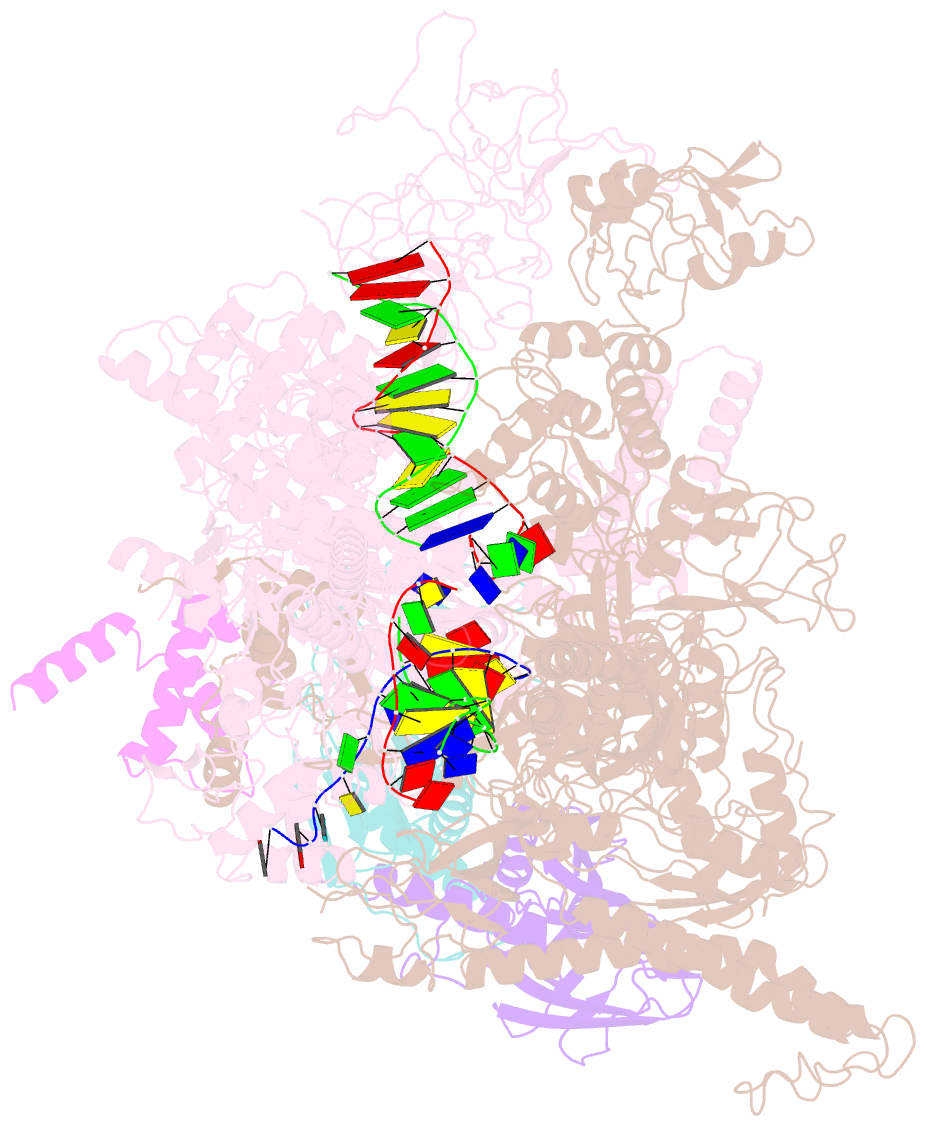
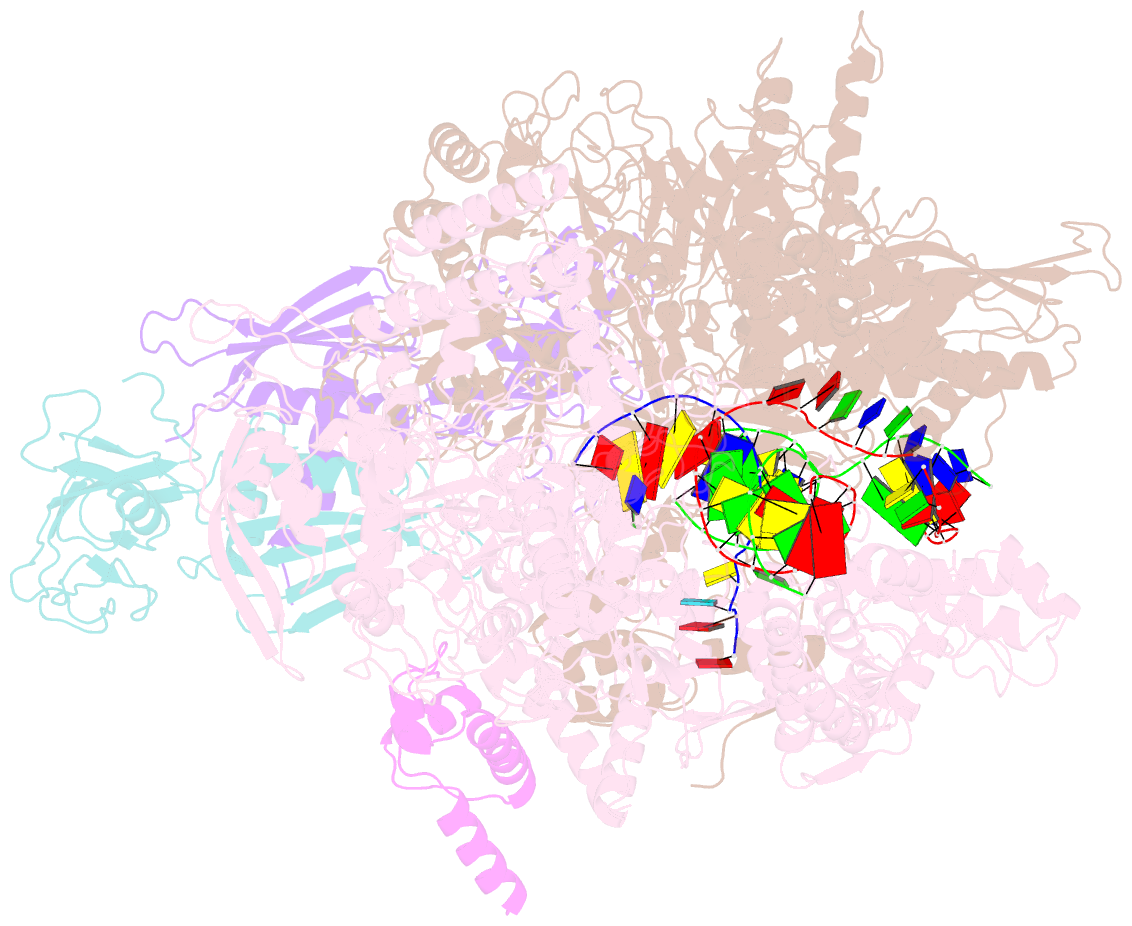
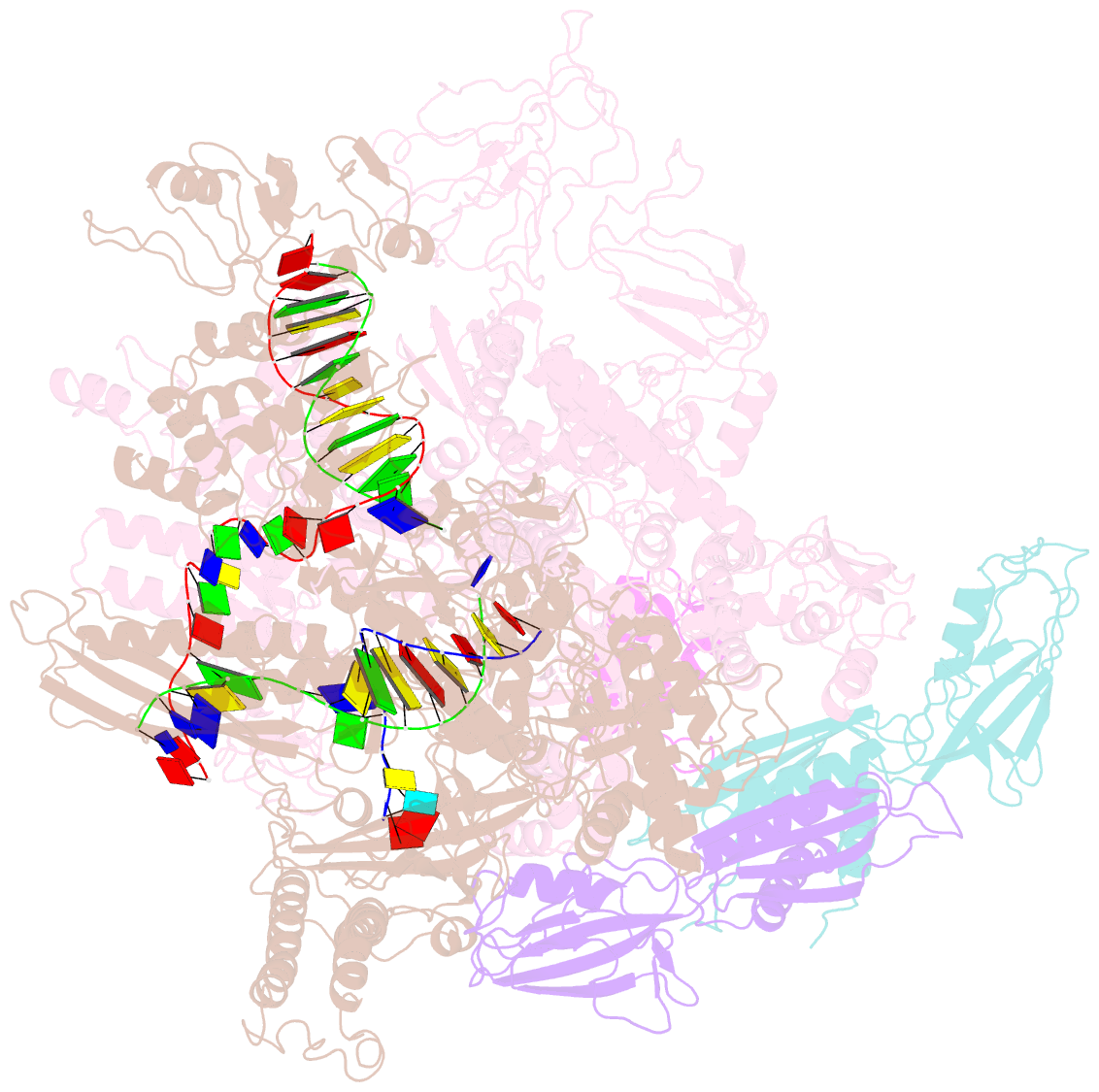
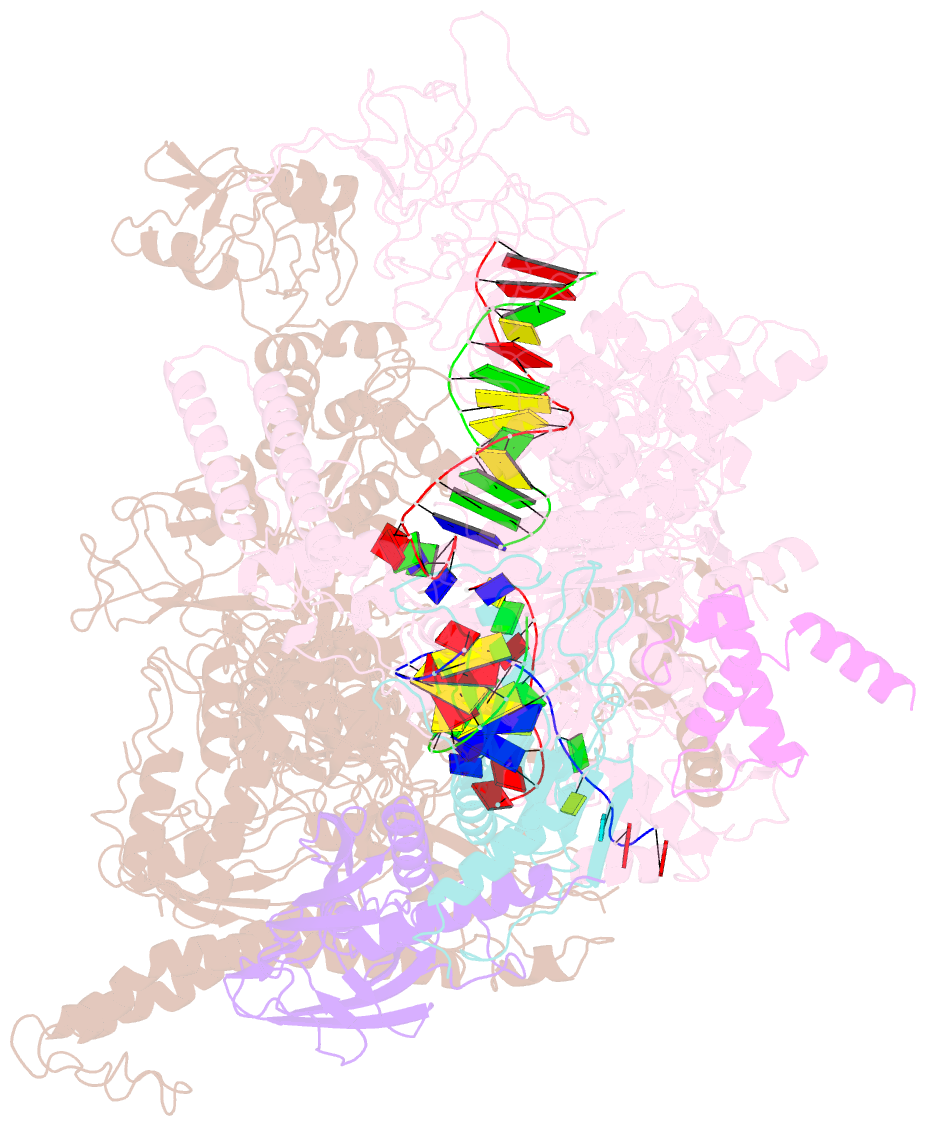
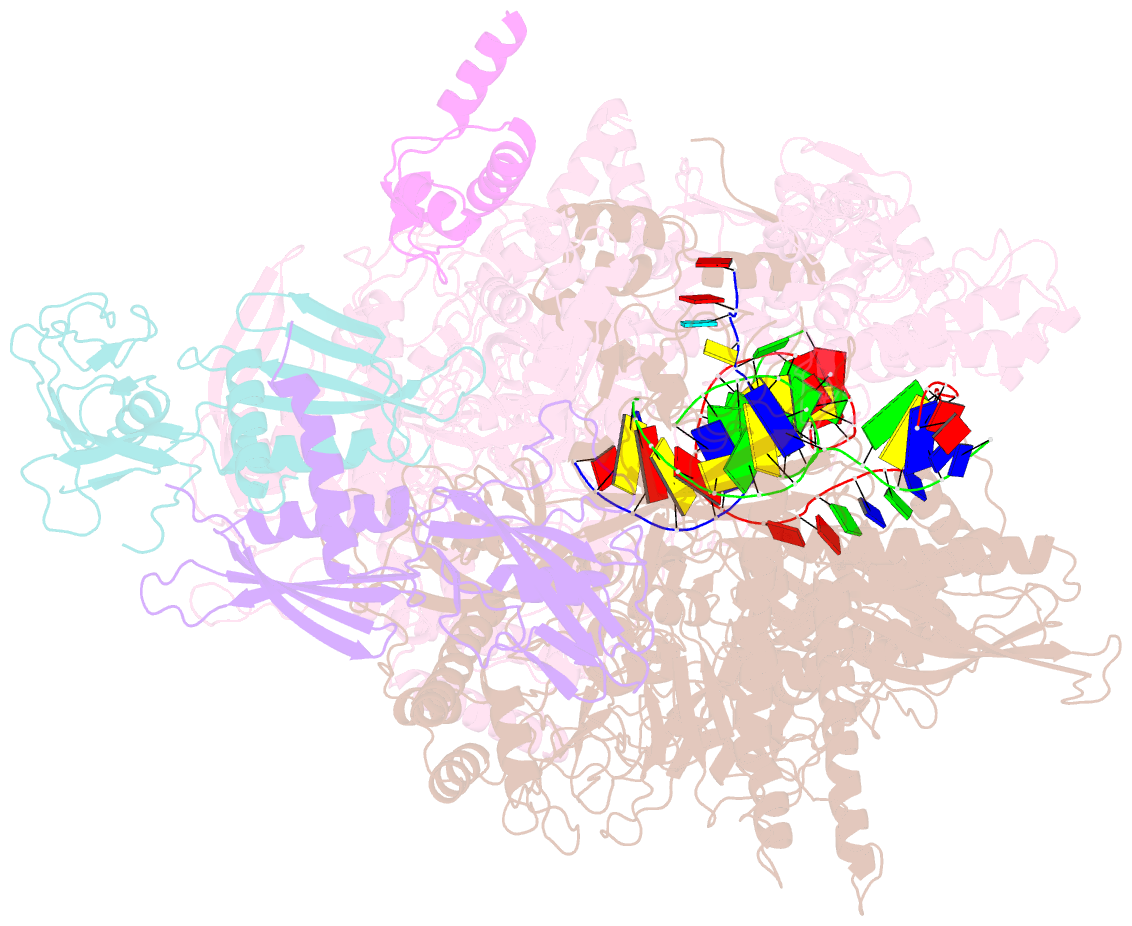
Summary information and primary citation
- PDB-id
- 8pbl; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- cryo-EM (2.87 Å)
- Summary
- E. coli RNA polymerase elongation complex stalled at thymine dimer lesion
- Reference
- Woodgate J, Mosaei H, Brazda P, Stevenson-Jones F, Zenkin N (2024): "Translation selectively destroys non-functional transcription complexes." Nature, 626, 891-896. doi: 10.1038/s41586-023-07014-3.
- Abstract
- Transcription elongation stalls at lesions in the DNA template1. For the DNA lesion to be repaired, the stalled transcription elongation complex (EC) has to be removed from the damaged site2. Here we show that translation, which is coupled to transcription in bacteria, actively dislodges stalled ECs from the damaged DNA template. By contrast, paused, but otherwise elongation-competent, ECs are not dislodged by the ribosome. Instead, they are helped back into processive elongation. We also show that the ribosome slows down when approaching paused, but not stalled, ECs. Our results indicate that coupled ribosomes functionally and kinetically discriminate between paused ECs and stalled ECs, ensuring the selective destruction of only the latter. This functional discrimination is controlled by the RNA polymerase's catalytic domain, the Trigger Loop. We show that the transcription-coupled DNA repair helicase UvrD, proposed to cause backtracking of stalled ECs3, does not interfere with ribosome-mediated dislodging. By contrast, the transcription-coupled DNA repair translocase Mfd4 acts synergistically with translation, and dislodges stalled ECs that were not destroyed by the ribosome. We also show that a coupled ribosome efficiently destroys misincorporated ECs that can cause conflicts with replication5. We propose that coupling to translation is an ancient and one of the main mechanisms of clearing non-functional ECs from the genome.