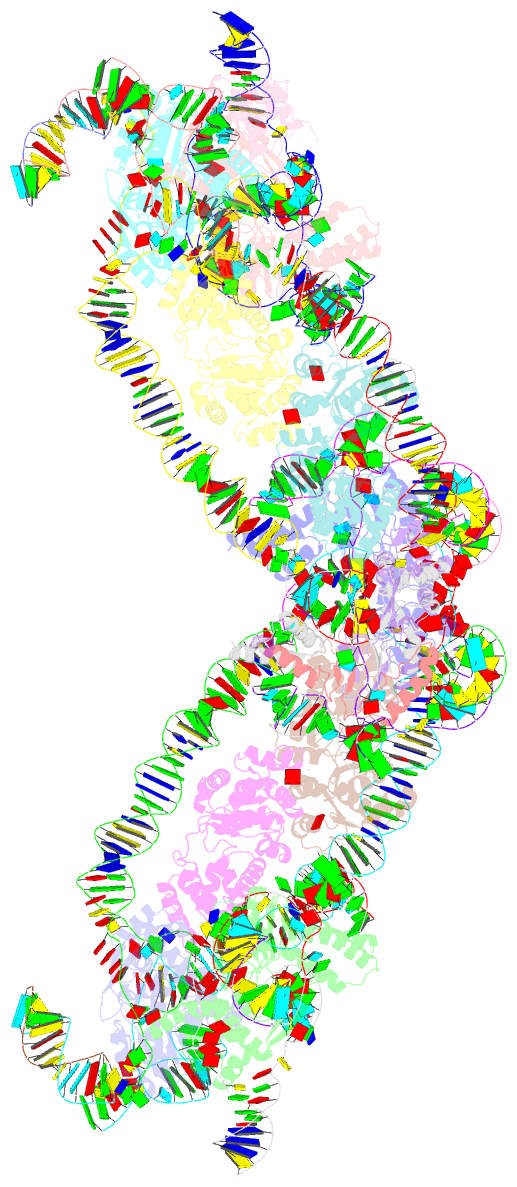
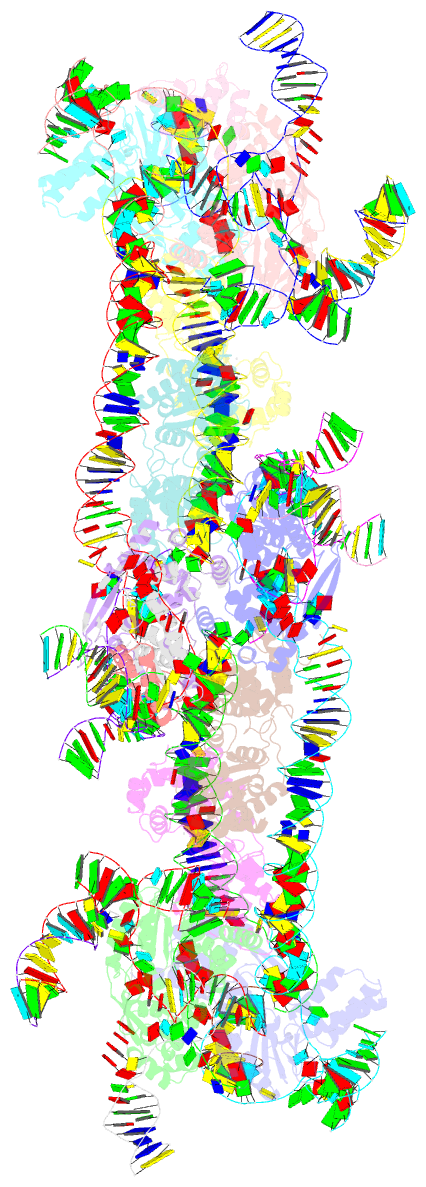
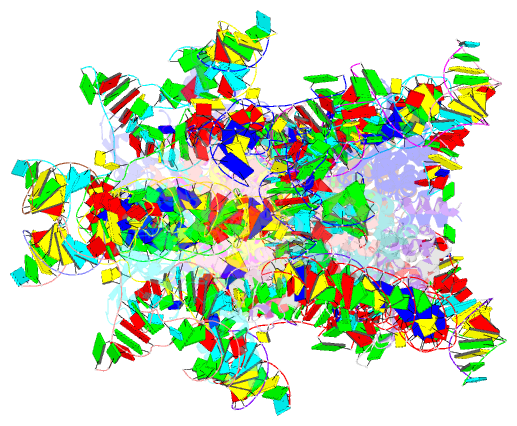
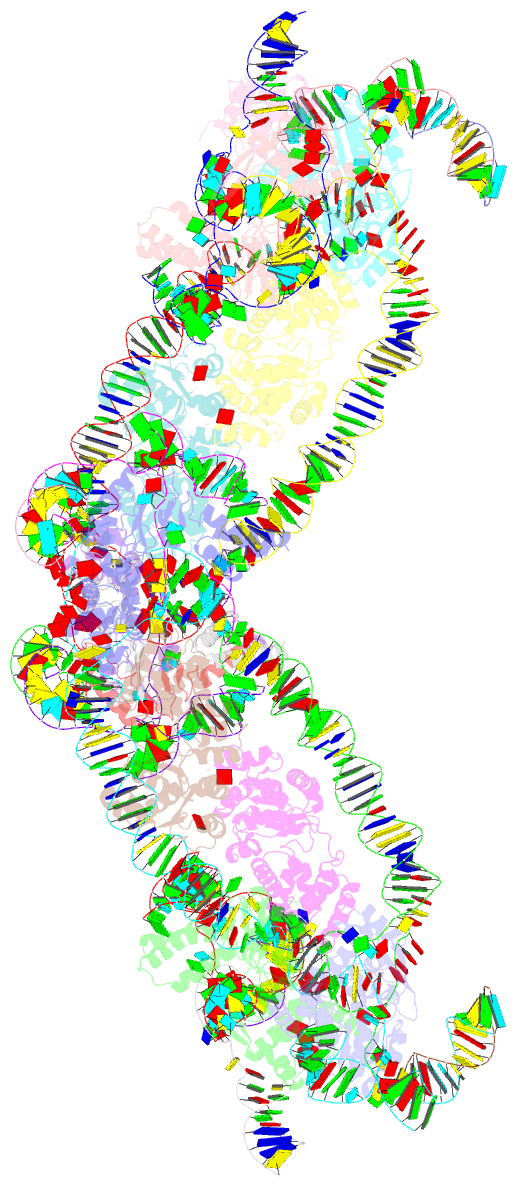
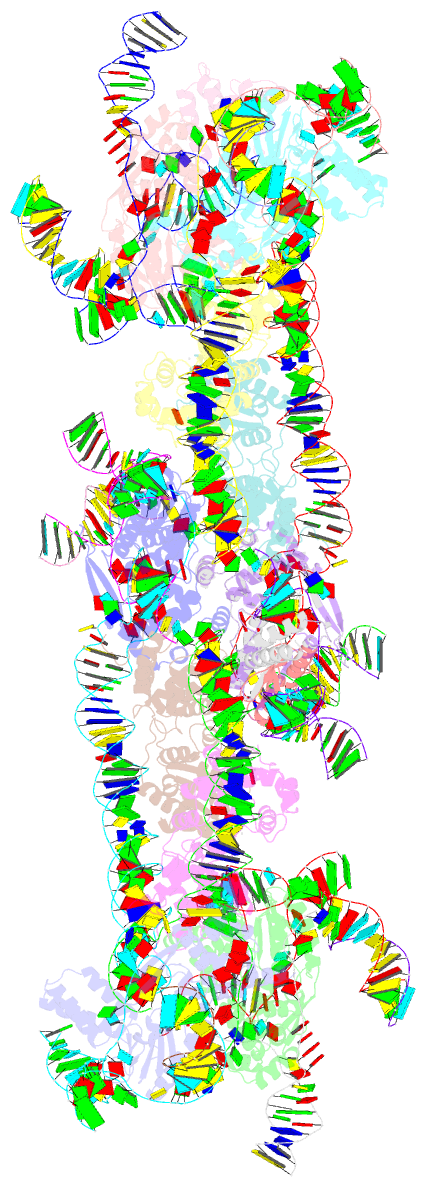
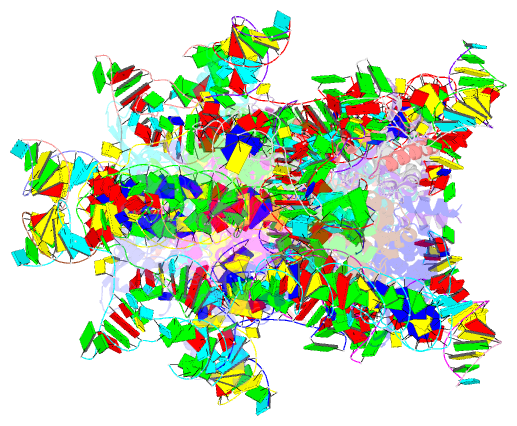
Summary information and primary citation
- PDB-id
- 8qbm; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- immune system
- Method
- cryo-EM (3.09 Å)
- Summary
- Retron-eco1 filament with adp-ribosylated effector (full map with 2 segments)
- Reference
- Carabias A, Camara-Wilpert S, Mestre MR, Lopez-Mendez B, Hendriks IA, Zhao R, Pape T, Fuglsang A, Luk SH, Nielsen ML, Pinilla-Redondo R, Montoya G (2024): "Retron-Eco1 assembles NAD + -hydrolyzing filaments that provide immunity against bacteriophages." Mol.Cell, 84, 2185. doi: 10.1016/j.molcel.2024.05.001.
- Abstract
- Retrons are toxin-antitoxin systems protecting bacteria against bacteriophages via abortive infection. The Retron-Eco1 antitoxin is formed by a reverse transcriptase (RT) and a non-coding RNA (ncRNA)/multi-copy single-stranded DNA (msDNA) hybrid that neutralizes an uncharacterized toxic effector. Yet, the molecular mechanisms underlying phage defense remain unknown. Here, we show that the N-glycosidase effector, which belongs to the STIR superfamily, hydrolyzes NAD+ during infection. Cryoelectron microscopy (cryo-EM) analysis shows that the msDNA stabilizes a filament that cages the effector in a low-activity state in which ADPr, a NAD+ hydrolysis product, is covalently linked to the catalytic E106 residue. Mutations shortening the msDNA induce filament disassembly and the effector's toxicity, underscoring the msDNA role in immunity. Furthermore, we discovered a phage-encoded Retron-Eco1 inhibitor (U56) that binds ADPr, highlighting the intricate interplay between retron systems and phage evolution. Our work outlines the structural basis of Retron-Eco1 defense, uncovering ADPr's pivotal role in immunity.