Summary information and primary citation
- PDB-id
- 8qqi; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- cryo-EM (2.9 Å)
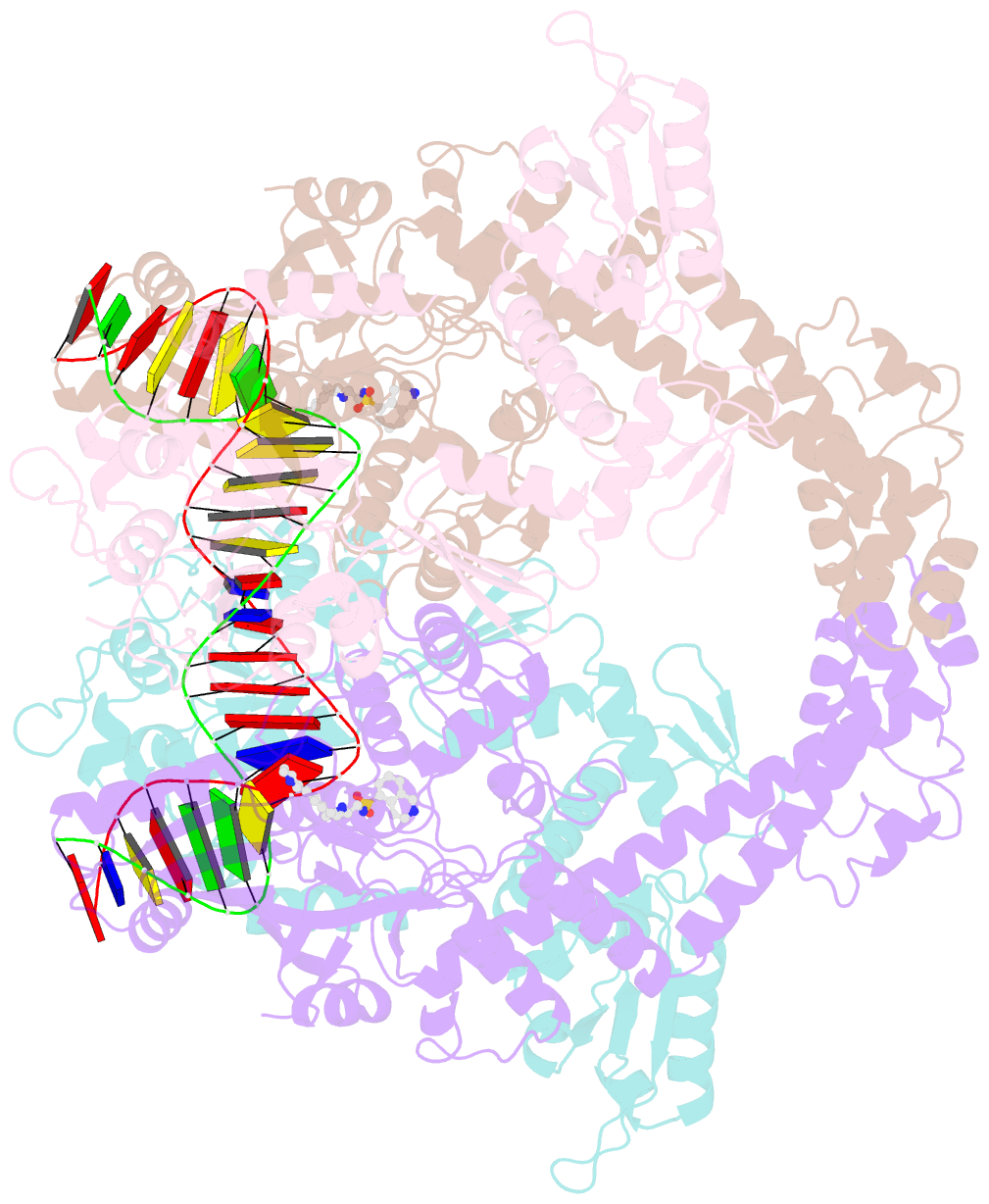
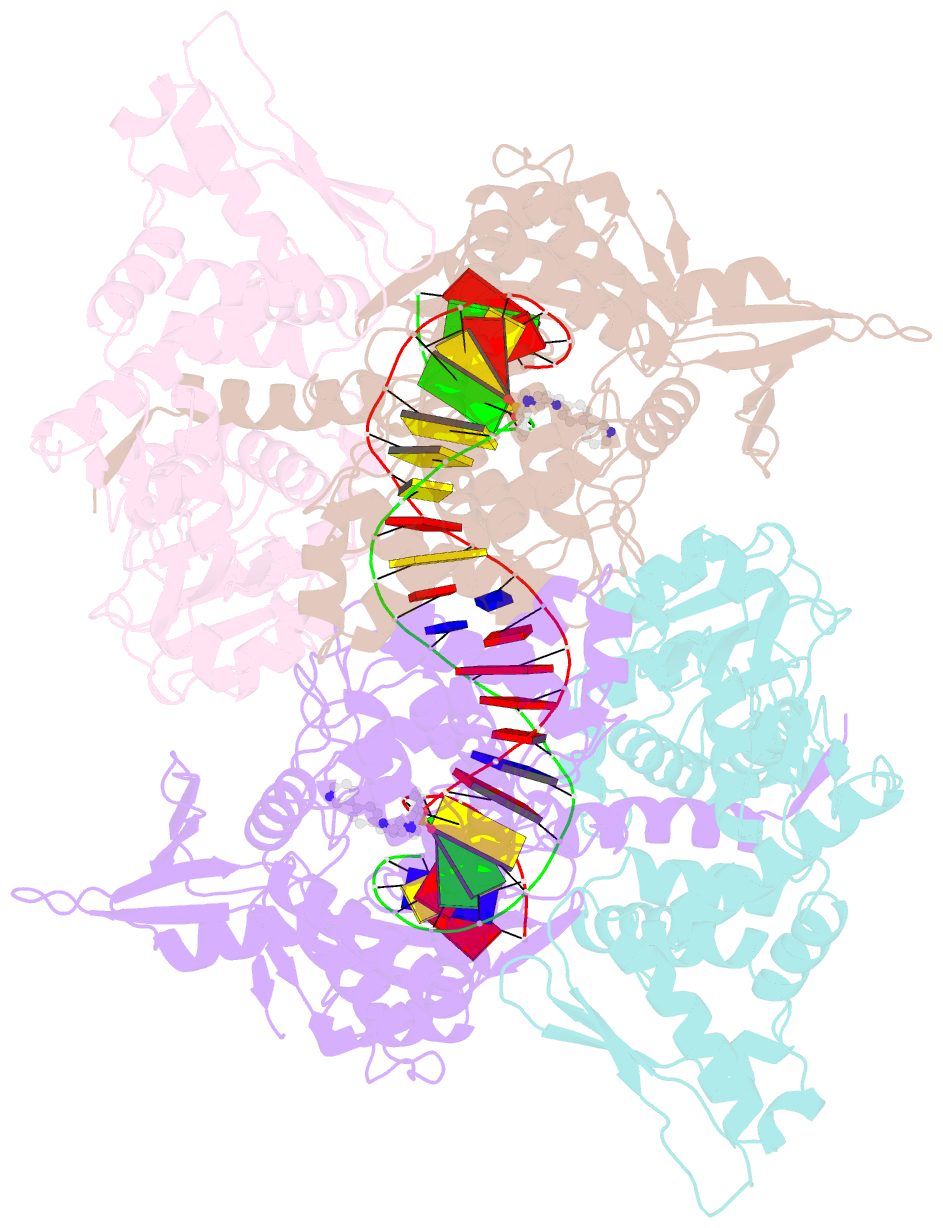
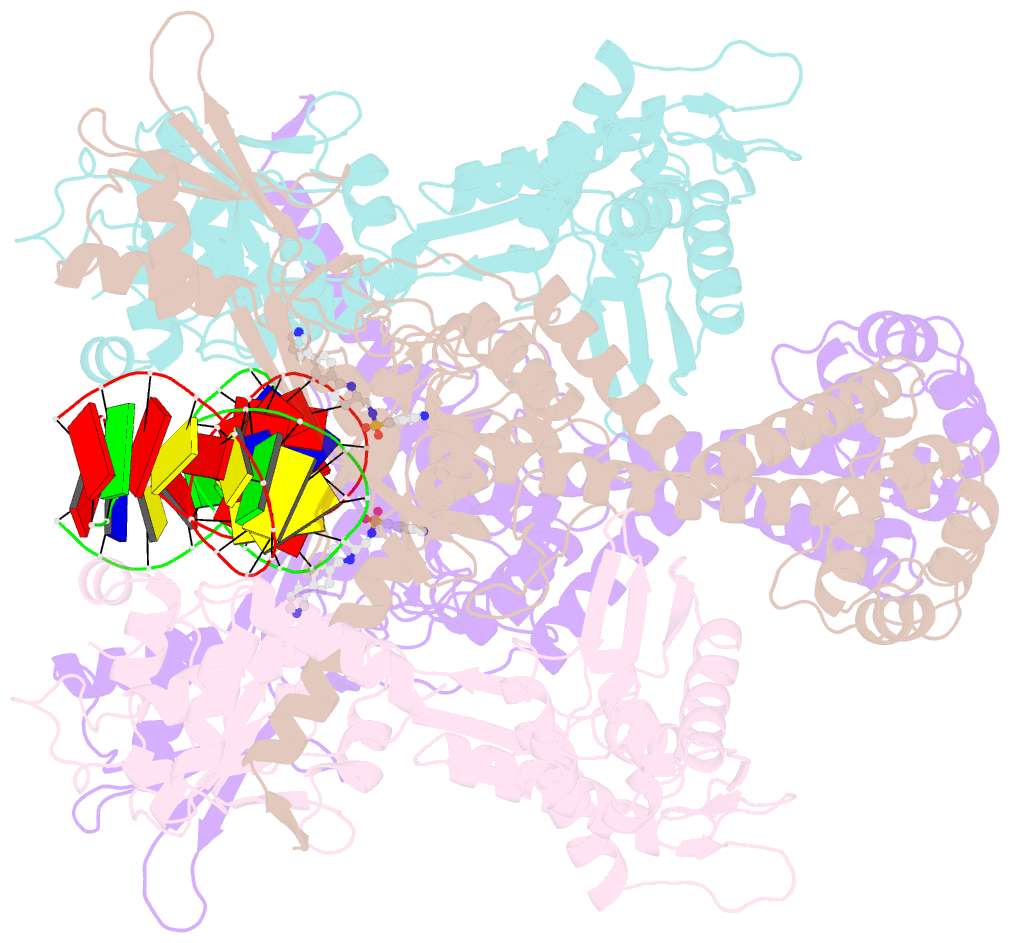
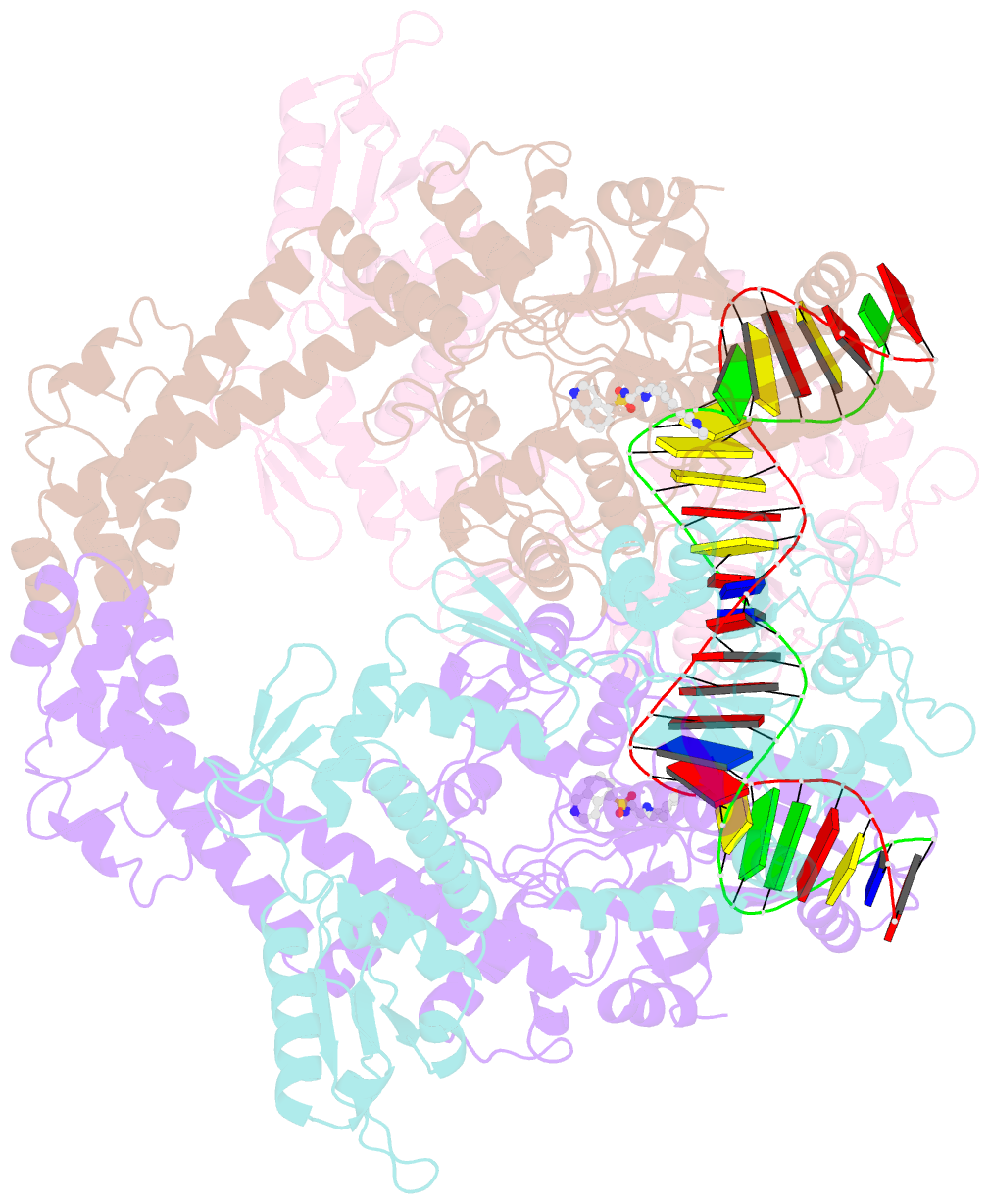
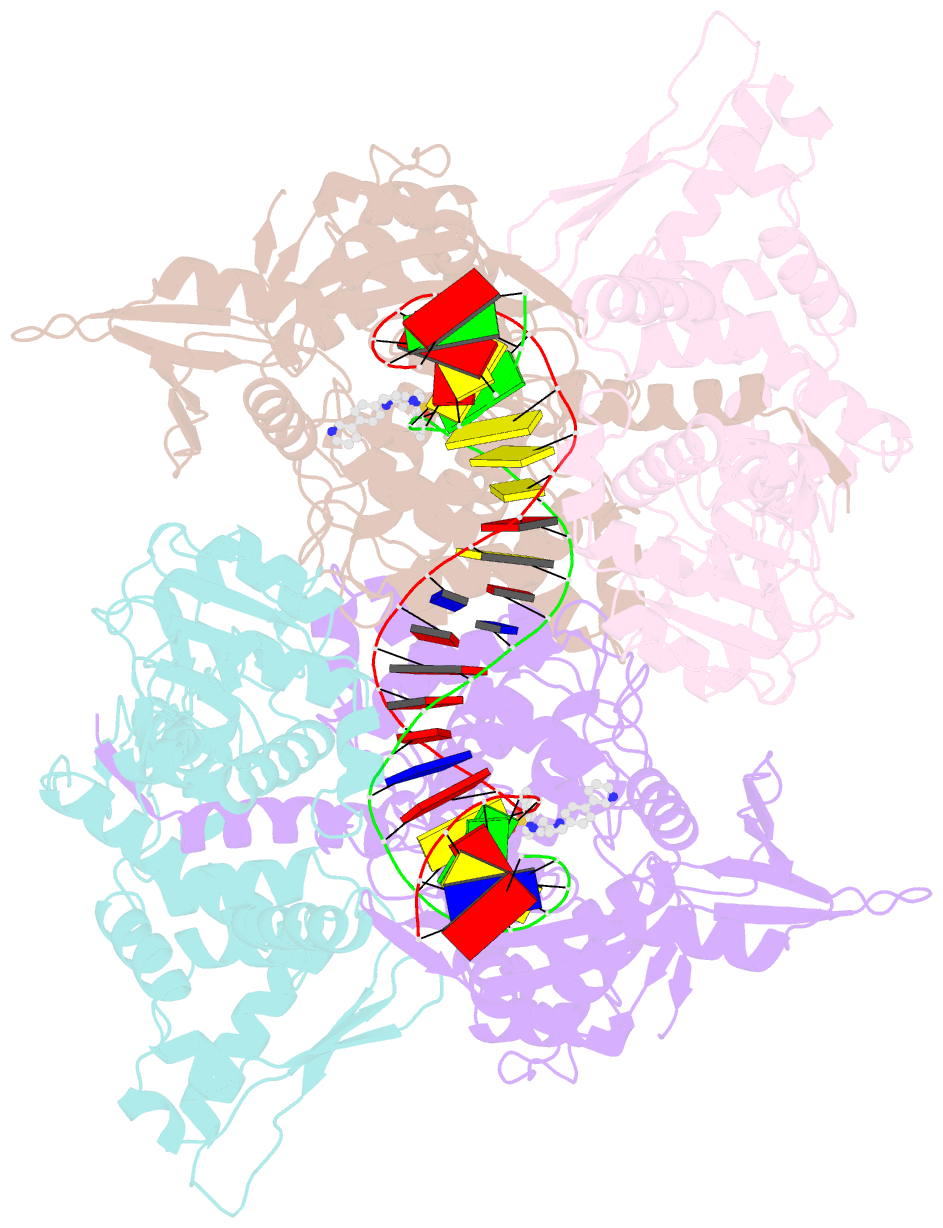
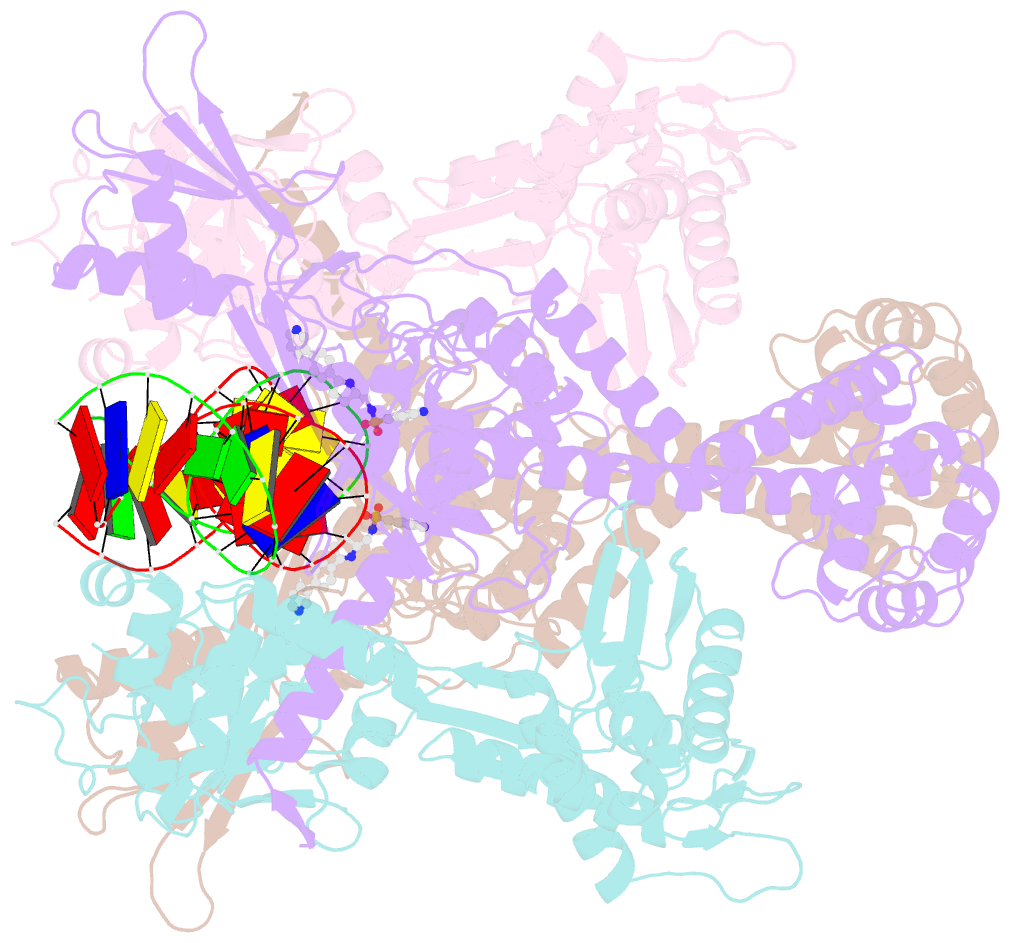
- Summary
- E.coli DNA gyrase in complex with 217 bp substrate DNA and lei-800
- Reference
- Bakker AT, Kotsogianni I, Avalos M, Punt JM, Liu B, Piermarini D, Gagestein B, Slingerland CJ, Zhang L, Willemse JJ, Ghimire LB, van den Berg RJHBN, Janssen APA, Ottenhoff THM, van Boeckel CAA, van Wezel GP, Ghilarov D, Martin NI, van der Stelt M (2024): "Discovery of isoquinoline sulfonamides as allosteric gyrase inhibitors with activity against fluoroquinolone-resistant bacteria." Nat.Chem., 16, 1462-1472. doi: 10.1038/s41557-024-01516-x.
- Abstract
- Bacteria have evolved resistance to nearly all known antibacterials, emphasizing the need to identify antibiotics that operate via novel mechanisms. Here we report a class of allosteric inhibitors of DNA gyrase with antibacterial activity against fluoroquinolone-resistant clinical isolates of Escherichia coli. Screening of a small-molecule library revealed an initial isoquinoline sulfonamide hit, which was optimized via medicinal chemistry efforts to afford the more potent antibacterial LEI-800. Target identification studies, including whole-genome sequencing of in vitro selected mutants with resistance to isoquinoline sulfonamides, unanimously pointed to the DNA gyrase complex, an essential bacterial topoisomerase and an established antibacterial target. Using single-particle cryogenic electron microscopy, we determined the structure of the gyrase-LEI-800-DNA complex. The compound occupies an allosteric, hydrophobic pocket in the GyrA subunit and has a mode of action that is distinct from the clinically used fluoroquinolones or any other gyrase inhibitor reported to date. LEI-800 provides a chemotype suitable for development to counter the increasingly widespread bacterial resistance to fluoroquinolones.