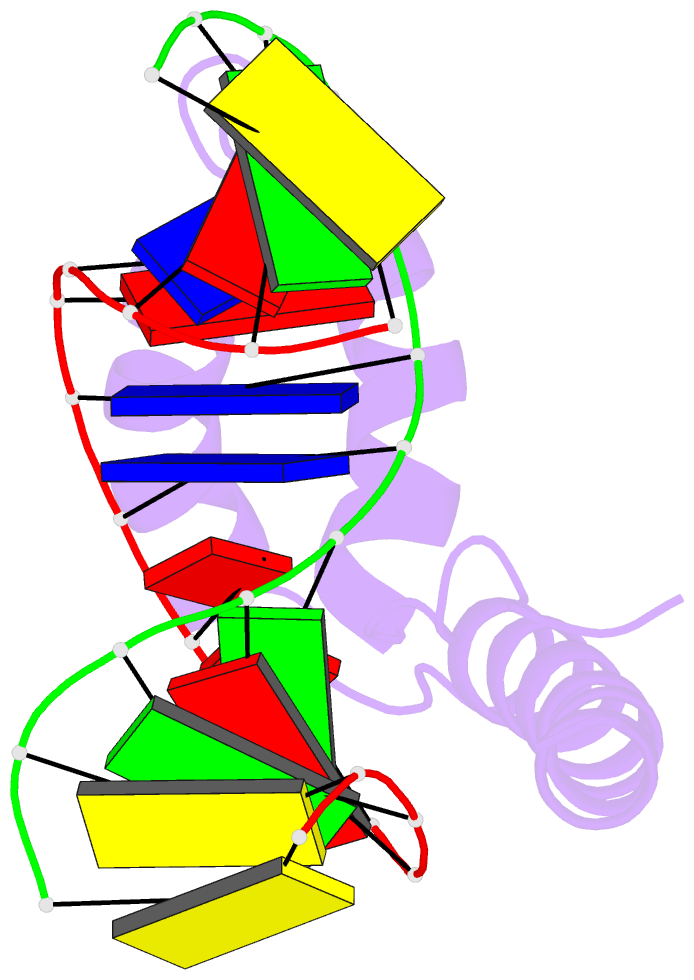
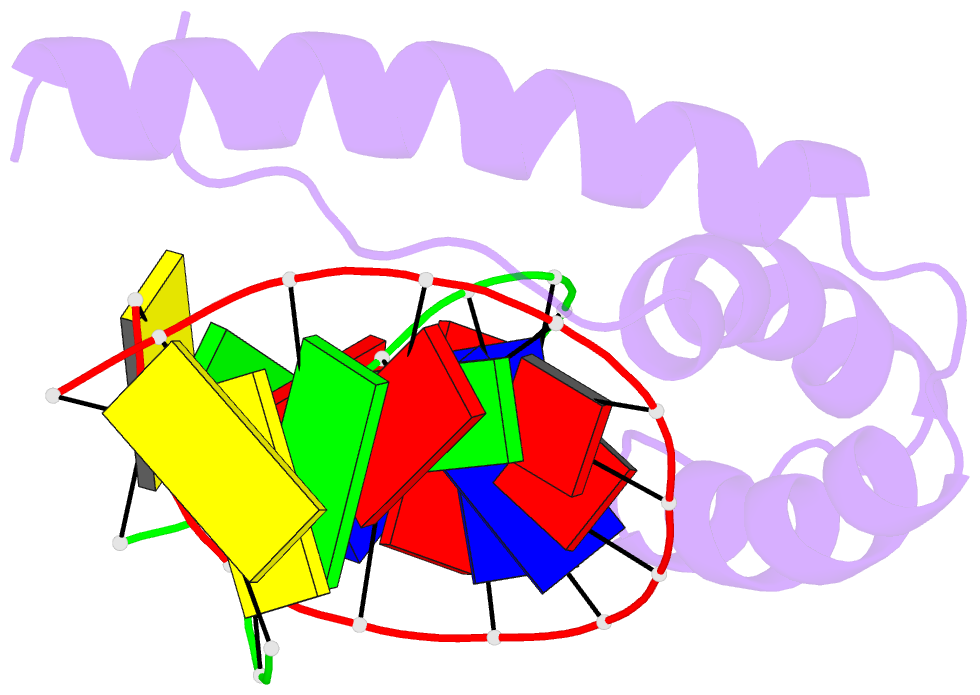
Summary information and primary citation
- PDB-id
- 8r1x; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- NMR
- Summary
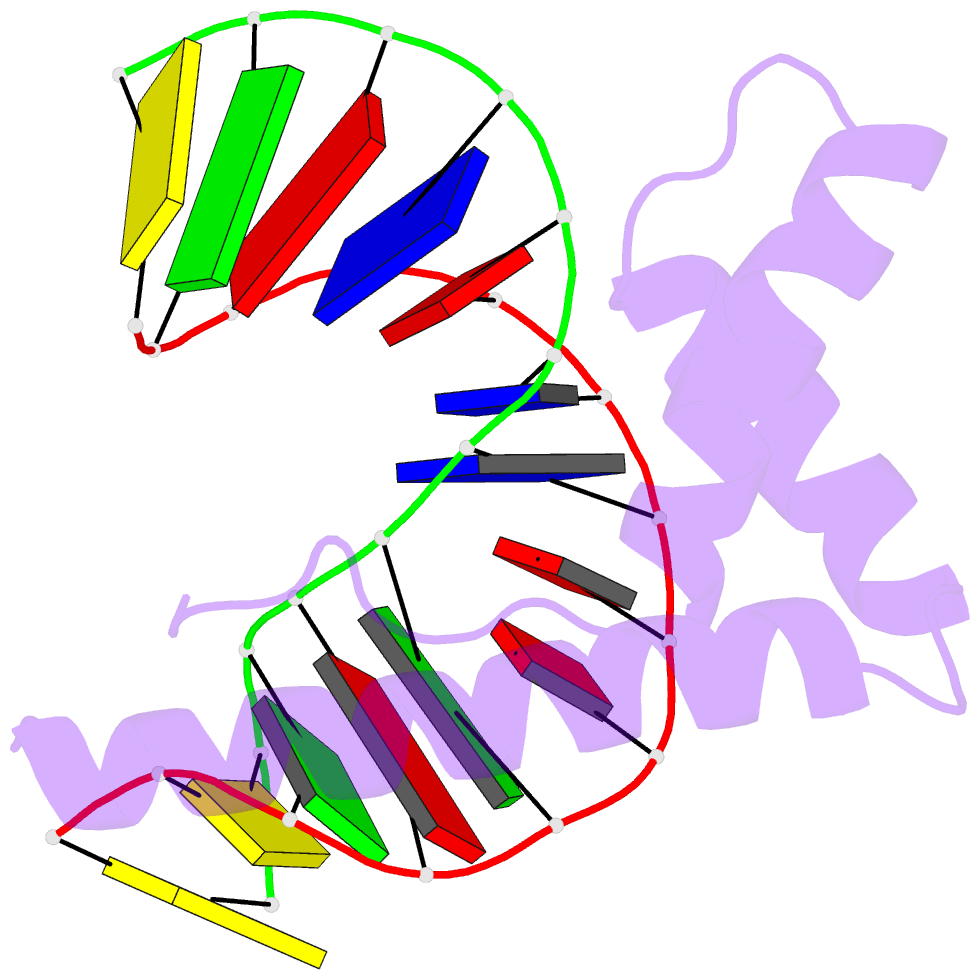
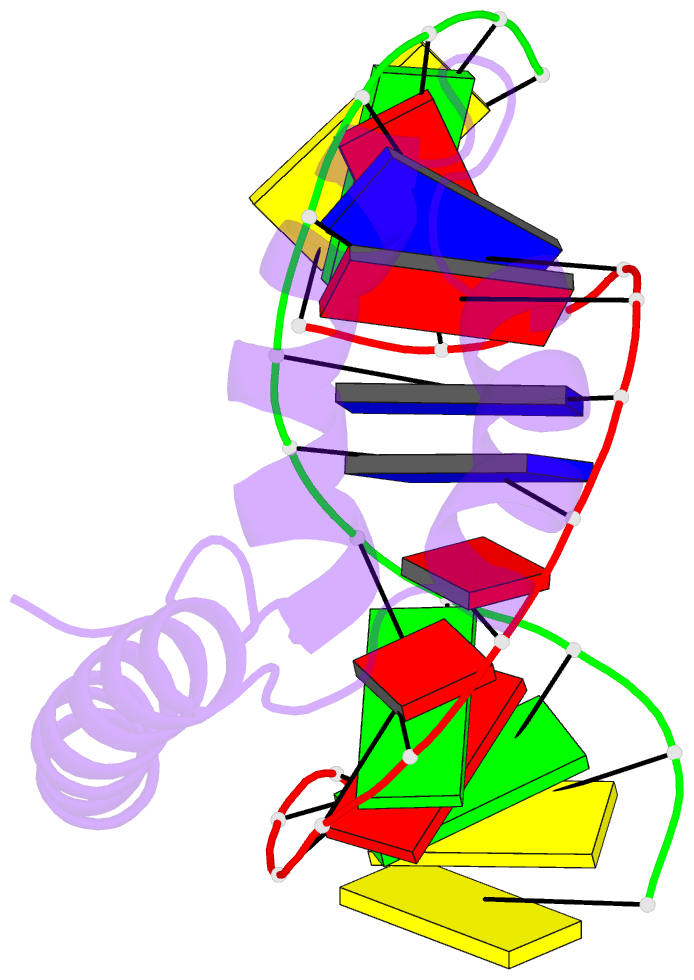
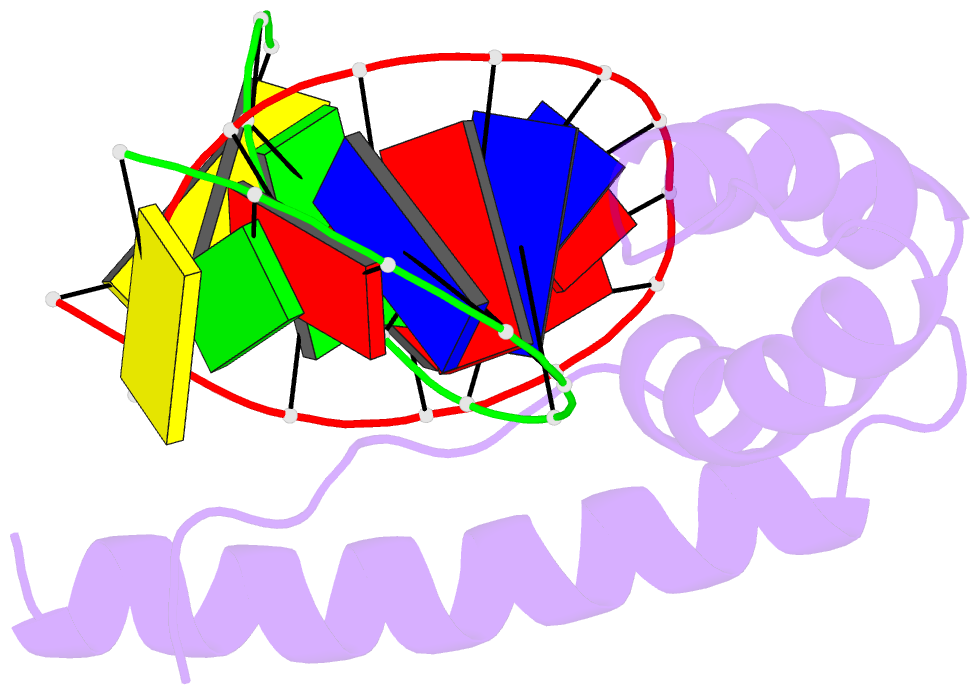
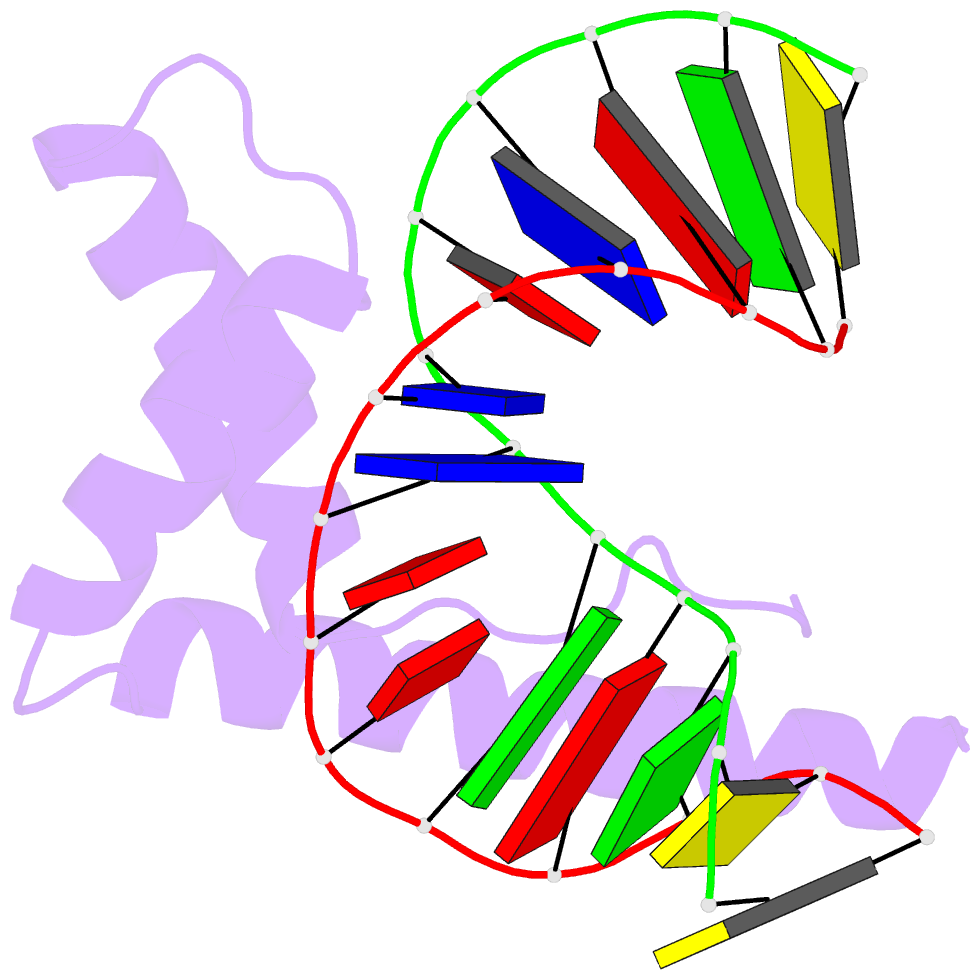
- Solution structure and chemical shift assignments for hmg-d y12f mutant complexed to a 14:12 da2 bulge DNA
- Reference
- Hill G, Yang JC, Easton L, Cerdan R, McLaughlin S, Stott K, Travers A, Neuhaus D (2024): "A Single Interfacial Point Mutation Rescues Solution Structure Determination of the Complex of HMG-D with a DNA Bulge." Chembiochem, e202400395. doi: 10.1002/cbic.202400395.
- Abstract
- Broadening of signals from atoms at interfaces can often be a limiting factor in applying solution NMR to the structure determination of complexes. Common contributors to such problems include exchange between free and bound states and the increased molecular weight of complexes relative to the free components, but another cause that can be more difficult to deal with occurs when conformational dynamics within the interface takes place at an intermediate rate on the chemical shift timescale. In this work we show how a carefully chosen mutation in the protein HMG-D rescued such a situation, making possible high-resolution structure determination of its complex with a dA2 bulge DNA ligand designed to mimic a natural DNA bend, and thereby revealing a new spatial organization of the complex.